Team:LMU-Munich/Spore Coat Proteins
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
Franzi.Duerr (Talk | contribs) |
||
| Line 10: | Line 10: | ||
| - | =='''Sporo'''beads - | + | =='''Sporo'''beads - What Protein Do ''You'' Want to Display?== |
<br> | <br> | ||
<div class="box"> | <div class="box"> | ||
| Line 33: | Line 33: | ||
<div class="box"> | <div class="box"> | ||
| - | ===Crust | + | ===Crust Protein Gene Organization=== |
{| "width=100%" style="text-align:center;" style="align:right"| | {| "width=100%" style="text-align:center;" style="align:right"| | ||
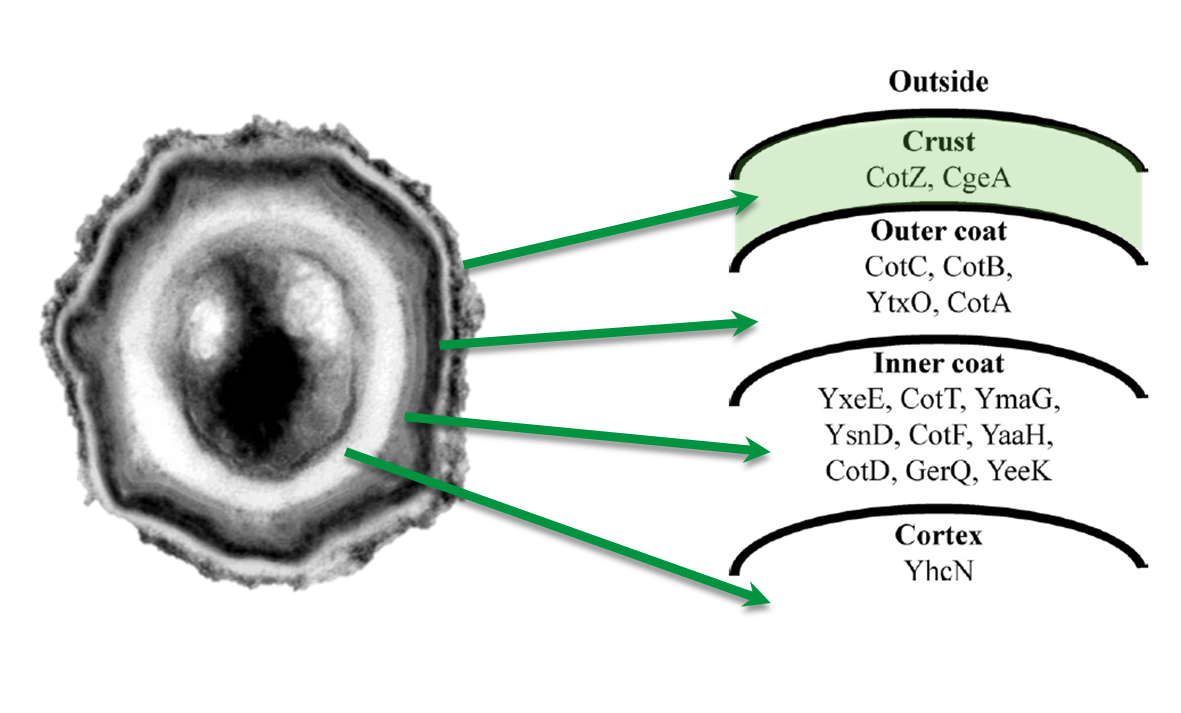
|<p align="justify">Location of the two crust protein genes in the genome</p> | |<p align="justify">Location of the two crust protein genes in the genome</p> | ||
| Line 42: | Line 42: | ||
</div> | </div> | ||
| - | < | + | <div class="box"> |
| - | + | ===Cloning Strategy=== | |
| - | + | {| "width=100%" style="text-align:center;" style="align:right"| | |
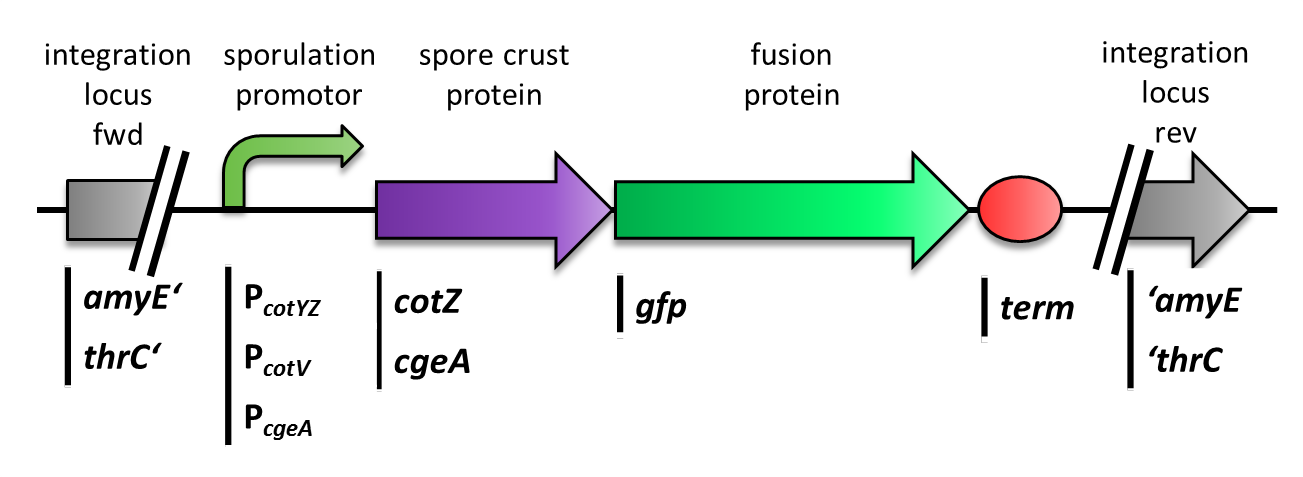
| - | {| | + | |<p align="justify">Cloning strategy to create different variants of our ''' Sporo'''beads</p> |
| - | + | |[[File:Final construct.png|200px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] | |
| - | + | ||
| - | |[[File: | + | |
|- | |- | ||
| - | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Spore_Coat_Proteins/cloning]] | |
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | </div> | |
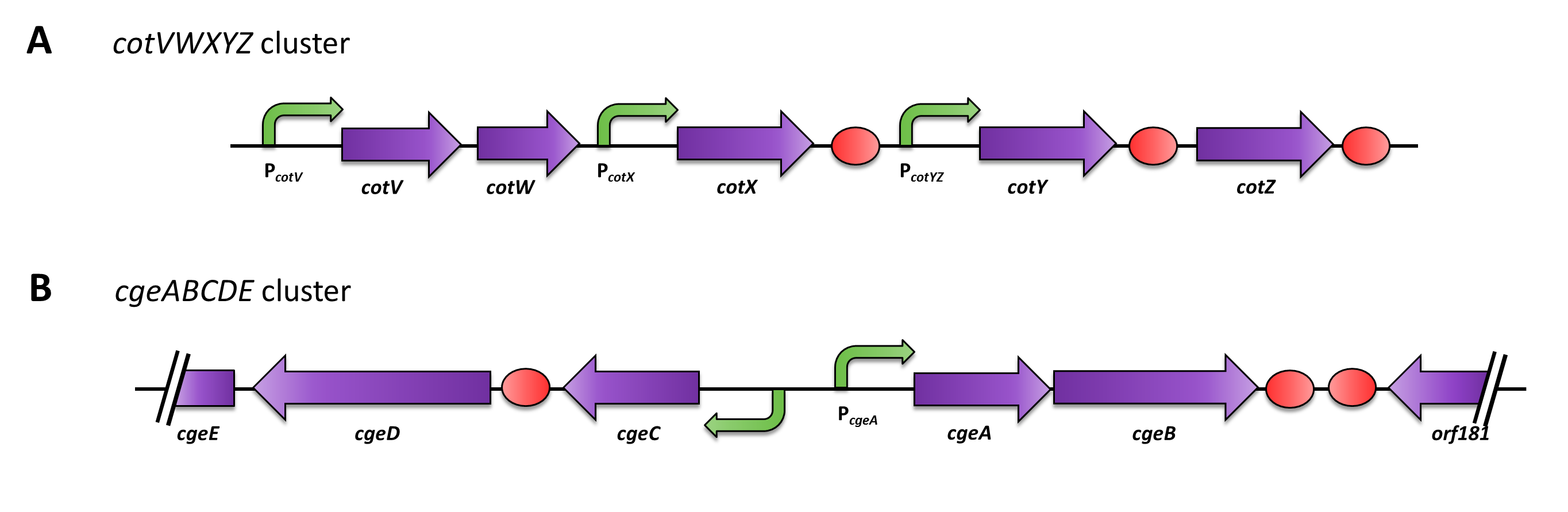
<p align="justify">Based on this data, two of the three promoters could be used for expression of spore crust fusion proteins (Fig. 3). First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> | <p align="justify">Based on this data, two of the three promoters could be used for expression of spore crust fusion proteins (Fig. 3). First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br>But we were able to finish five constructs and integrated them into wild type W168 and the Δ''cotZ'' mutant: | <br>But we were able to finish five constructs and integrated them into wild type W168 and the Δ''cotZ'' mutant: | ||
</p> | </p> | ||
| Line 190: | Line 153: | ||
|} | |} | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | [[File:Spore crust proteins cycle.jpg|600px|right]] | ||
| + | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | | style="width: 100%;background-color: #FFFFFF;" | | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #FFFFFF;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #FFFFFF;" | | ||
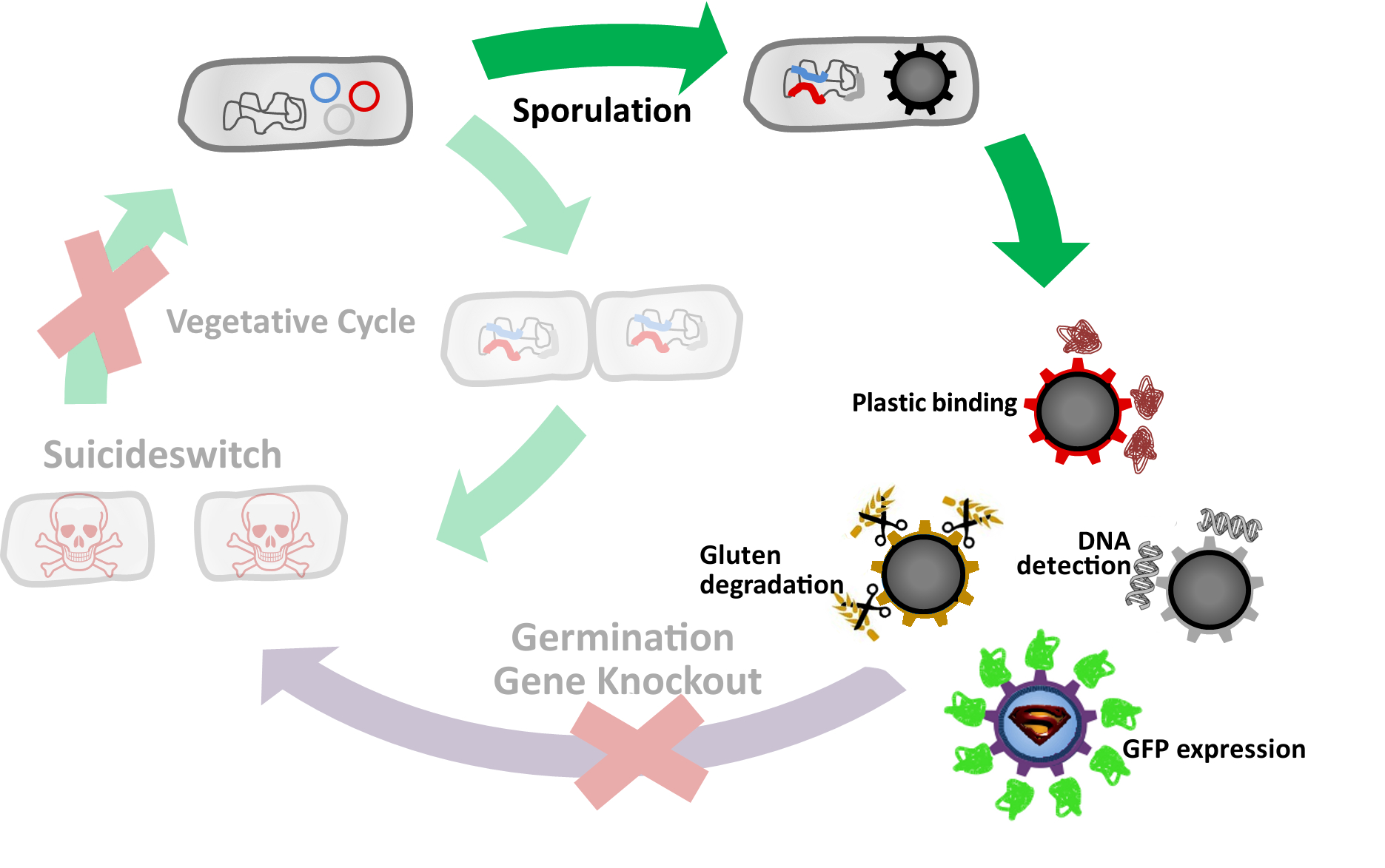
| + | <font color="#000000"; size="2">Fig. 3: The '''Bead'''zillus cycle</font> | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | |||
<!-- Include the next line at the end of every page --> | <!-- Include the next line at the end of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Revision as of 12:29, 23 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Sporobeads - What Protein Do You Want to Display?
Crust Promoter Evaluation
Three sporulation-dependent promoters (PcotYZ, PcotV, PcgeA) were constructed, two of which show the expected behavior. | 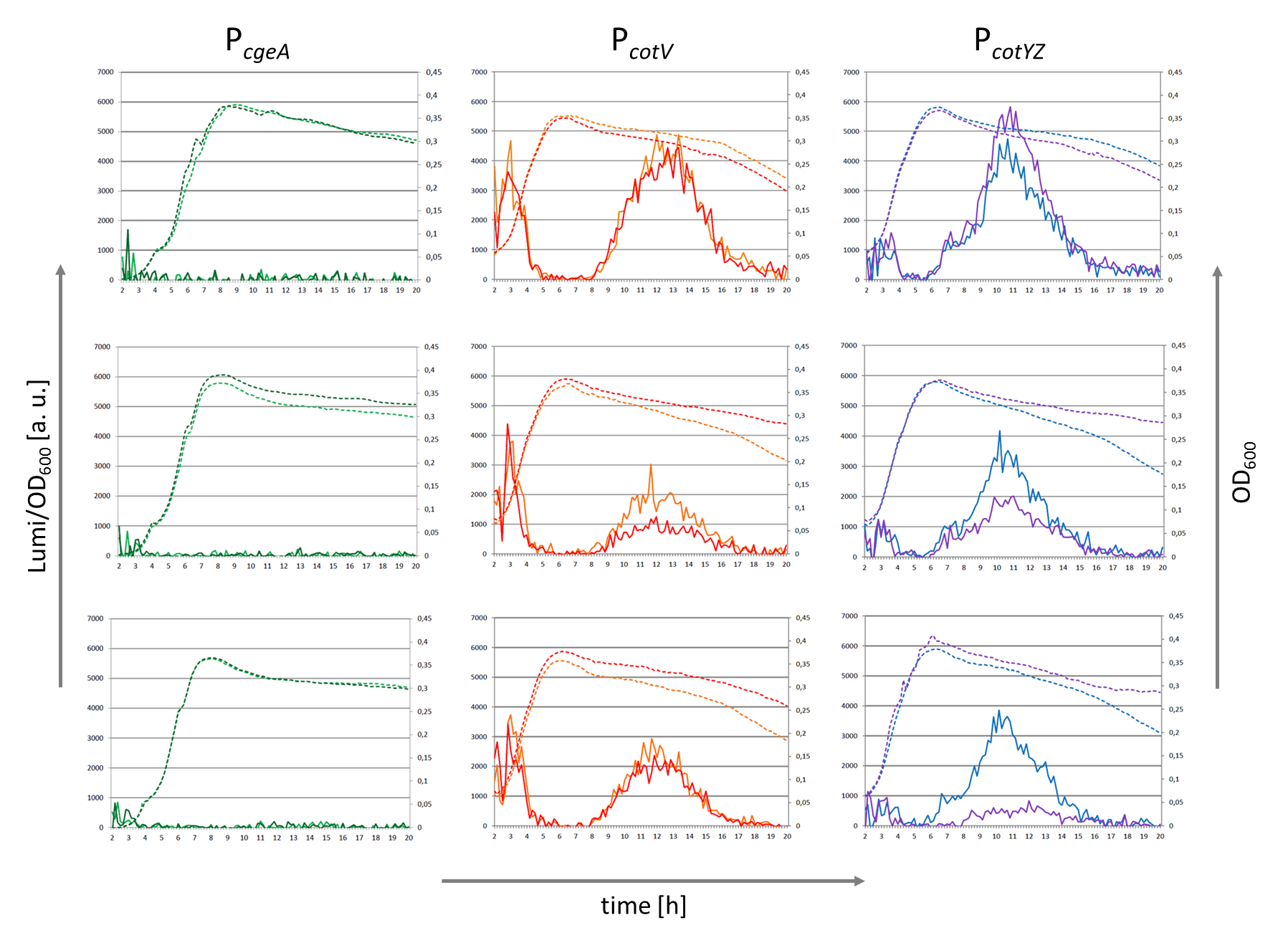
|
Based on this data, two of the three promoters could be used for expression of spore crust fusion proteins (Fig. 3). First, we used gfp as a proof of principle and fused it to cgeA and cotZ. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.
But we were able to finish five constructs and integrated them into wild type W168 and the ΔcotZ mutant:
</p>
| recipient strain W168 | recipient strain B 49 (W168 ΔcotZ) | |
|---|---|---|
| pSBBs1C-PcotYZ-cotZ-2aa-gfp-terminator | B 53 | B 70 |
| pSBBs1C-PcotYZ-cotZ-gfp-terminator | B 54 | B 71 |
| pSBBs1C-PcotV-cotZ-2aa-terminator | B 55 | B 72 |
| pSBBs1C-PcotV-cotZ-terminator | B 56 | B 73 |
| pSBBs1C-PcgeA-cotZ-2aa-terminator | B 52 | B 69 |
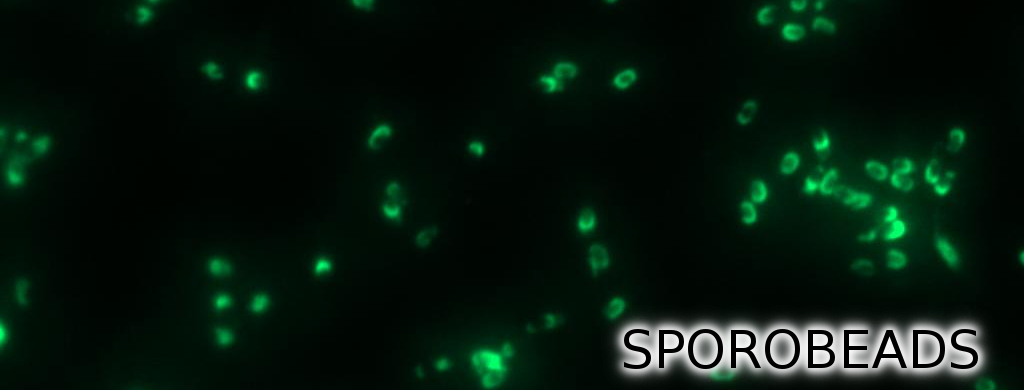
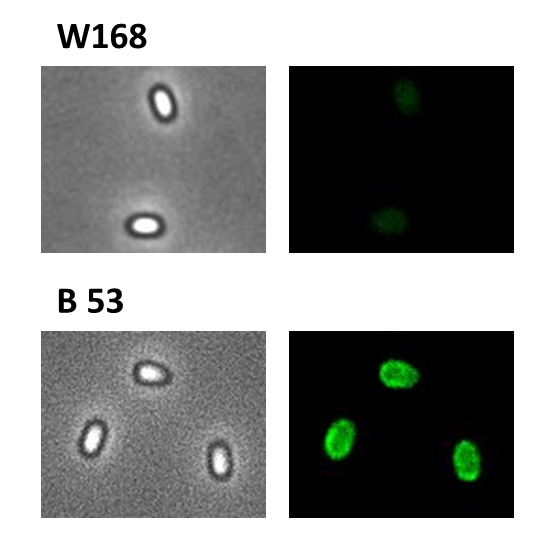
Finally, we started with the most important experiment for our GFP-Sporobeads, the fluorescence microscopy. We developed a sporulation protocol (for details see Protocol for enhancement of mature spore numbers) that increases the rates of mature spores in our samples. The cells were fixed on agarose pads and investigated by phase contrast and fluorescence microscopy. While spores of the wild type only showed the known background fluorescence, all Sporobeads showed bright green fluorescence at the edge (on the surface) of the spores. Sporobeads from strain B 53 (containing the PcotYZ-cotZ-2aa-gfp-terminator construct) showed the highest fluorescence intensity (see Fig. 5 and all data). Hence, this strain was chosen for further experiments.
|
Since there were still some vegetative cells left after 24 hours of growth in Difco Sporulation Medium, we wanted to purify the Sporobeads. We tried three different methods for this approach: treatment with French Press, sonification and lysozyme. By means of the microscopy results we were able to conclude that lysozyme treatment was the only successful method (see data). Additionally, this treatment did not harm the crust fusion proteins as green fluorescence was still detectable afterwards (see data for details).
Moreover, we wondered if clean deletions of the native spore crust genes would show any difference in fusion protein expression in our Sporobeads. Thus, we deleted the native cotZ and cgeA using the pMAD based gene deletion strategy described by Arnaud et al., 2004.
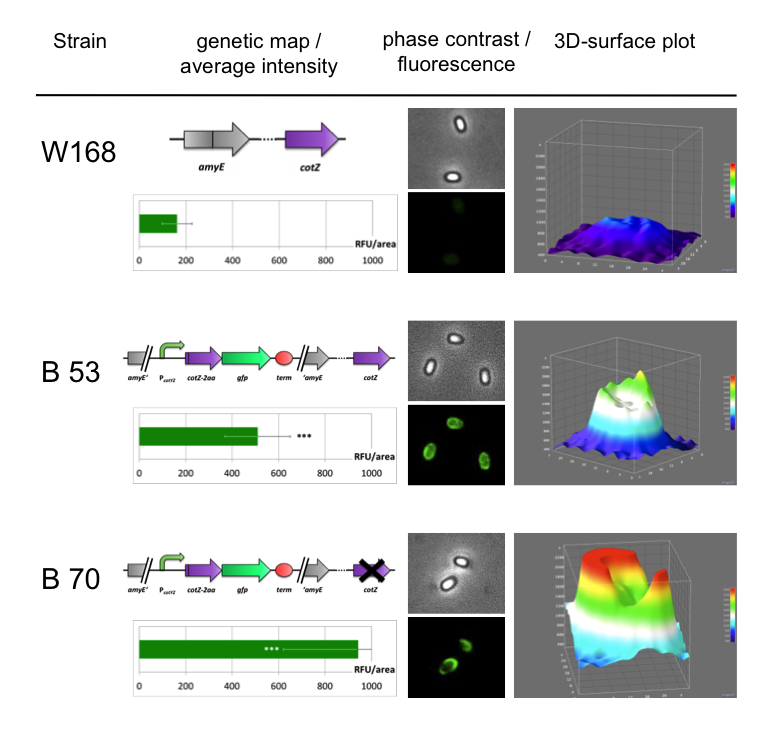
Because of the low but distinct fluorescence of wild type spores, we measured and compared the fluorescence intensity of 100 spores per construct (see data). We obtained significant differences between wild type spores and all of our Sporobeads (see data). The intensity bar charts in Fig. 6 show the fluorescence intensity, while the 3D graphs illustrate the distribution of fluorescence intensity across the spore surface. This correlates with the localization of our fusion proteins in the crust. For image analysis we measured the fluorescence intensity of an area of 750 pixel per spore by using ImageJ and evaluated the results with the statistical software R. The following graph (Fig. 6) shows the results of microscopy and ImageJ analysis of the strongest construct integrated into wildtype W168 (B53) and the deletion strain B 49 (B70).
|
| |
|
As shown in Fig. 6, the wild type spore has hardly any fluorescence, whereas both Sporobeads with the integrated construct pSBBs1C-PcotYZ-cotZ-2aa-gfp-terminator give a distinct fluorescence signal around the edge of the spore. Furthermore, it demonstrates that strain B 70 has the highest fluorescence intensity.
In summary we successfully developed functional sporobeads that are capable of displaying any protein of choice on the surface of modified B. subtilis endospores.

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
|
 "
"