Team:LMU-Munich/Results
From 2012.igem.org
| (21 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
| - | ===''B. subtilis'' | + | ===Seven novel vectors were constructed and four already proven to work in ''B. subtilis''.=== |
| + | <br> | ||
{| class="colored" | {| class="colored" | ||
!Name | !Name | ||
| Line 83: | Line 84: | ||
| - | + | ===15 Promoters evaluated in ''B. subtilis'' and added to the registry!=== | |
| - | + | <br> | |
| - | === | + | <p align="justify"> This section gives an overview on the strength of all evaluated promoters, which span a large range of activities. For more details and informations of the experiments see the [https://2012.igem.org/Team:LMU-Munich/Data Data] page of the promoters. Note that P<sub>''veg''</sub> was not evaluated with luminescence measurements and this bar is just projected from the results of the β-galactosidase assay.</p> |
| - | + | ||
| - | <p align="justify"> This section gives an overview of all evaluated promoters which | + | |
<br> | <br> | ||
| - | |||
{| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
| Line 98: | Line 96: | ||
{| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
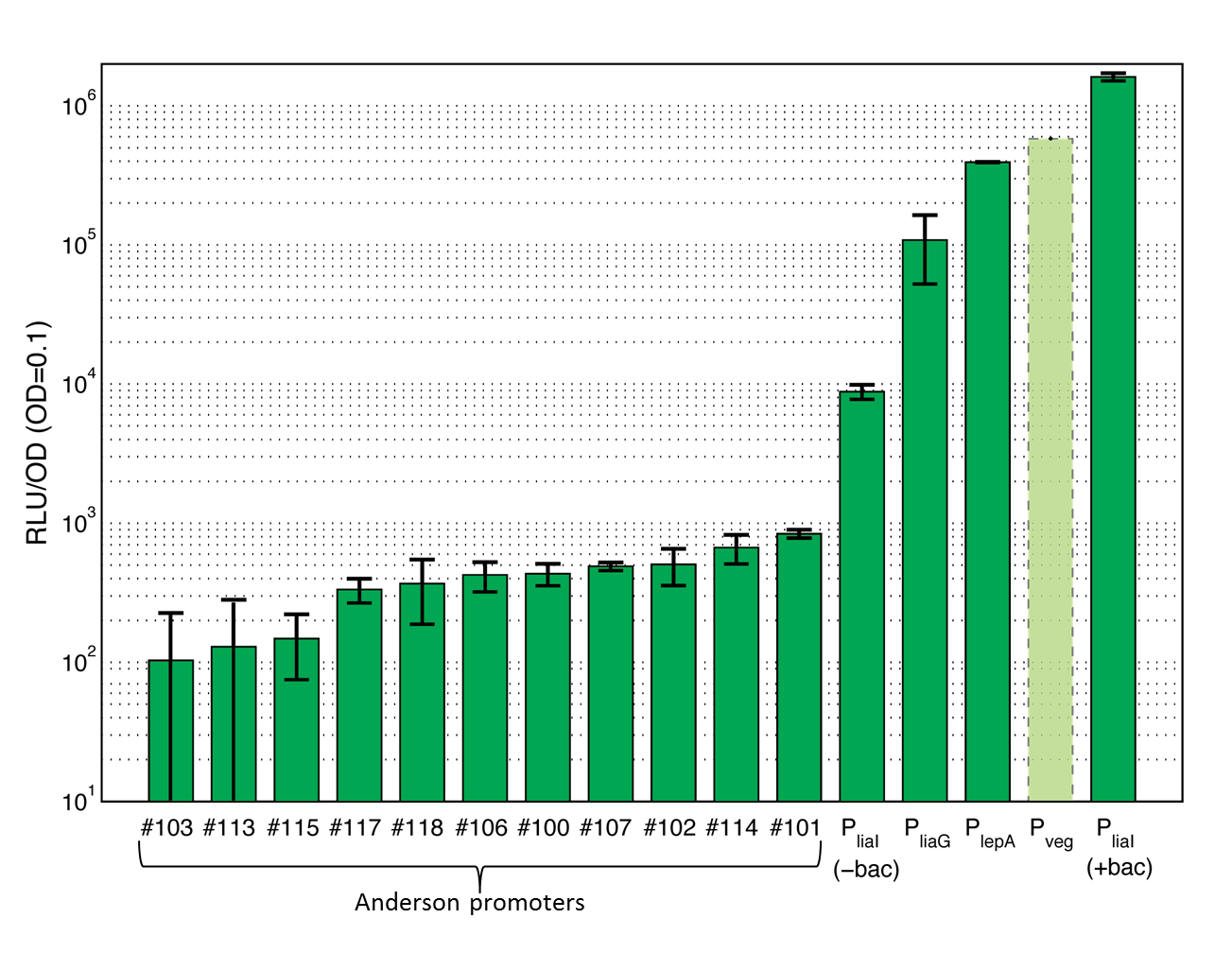
| - | <font color="#000000"; size="2"><p align="justify"> '''Overview of promoter activity evaluated with luminescence measurements in pSB<sub>''Bs''</sub>3C-''luxABCDE''.''' These values derive from the experiments you can find in our Data section. Lumi per OD<sub>600</sub> are taken at a OD<sub>600</sub> of 0.1. Values are the average and the standard deviation of three different experiments for clone 1. Shown is the activity of the Anderson promoters J23100 (#100), J23101 (#101), J23102 (#102), J23103 (#103), J23106 (#106), J23107 (#107), J23113 (#113), J23114 (#114), J23115 (#115), J23117 (#117), J23118 (#118) as well as the activity of the constitutive promoters P<sub>''liaG''</sub>, and P<sub>''lepA''</sub>. The activity of the inducible promoter P<sub>''liaI''</sub> is shown with (+bac) and without (-bac) induction with bacitracin (10 μg/ml). The promoter activity of P<sub>''veg''</sub> is projected from the results from the | + | <font color="#000000"; size="2"><p align="justify"> '''Overview of promoter activity evaluated with luminescence measurements in pSB<sub>''Bs''</sub>3C-''luxABCDE''.''' These values derive from the experiments you can find in our Data section. Lumi per OD<sub>600</sub> are taken at a OD<sub>600</sub> of 0.1. Values are the average and the standard deviation of three different experiments for clone 1. Shown is the activity of the Anderson promoters J23100 (#100), J23101 (#101), J23102 (#102), J23103 (#103), J23106 (#106), J23107 (#107), J23113 (#113), J23114 (#114), J23115 (#115), J23117 (#117), J23118 (#118) as well as the activity of the constitutive promoters P<sub>''liaG''</sub>, and P<sub>''lepA''</sub>. The activity of the inducible promoter P<sub>''liaI''</sub> is shown with (+bac) and without (-bac) induction with bacitracin (10 μg/ml). The promoter activity of P<sub>''veg''</sub> is projected from the results from the β-galactosidase assay and was not measured with luminescence measurements. For the assays shown, an intermediate version of the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' was used, which still contained one forbidden PstI site.</p></font> |
|} | |} | ||
|} | |} | ||
| Line 104: | Line 102: | ||
| + | ===Sporobeads - what protein do you want to display?=== | ||
| + | <br> | ||
| + | {| style="color:black;" cellpadding="3" width="100%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:Fluorescence of Sporobeads.png|610px|center]] | ||
| + | |} | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
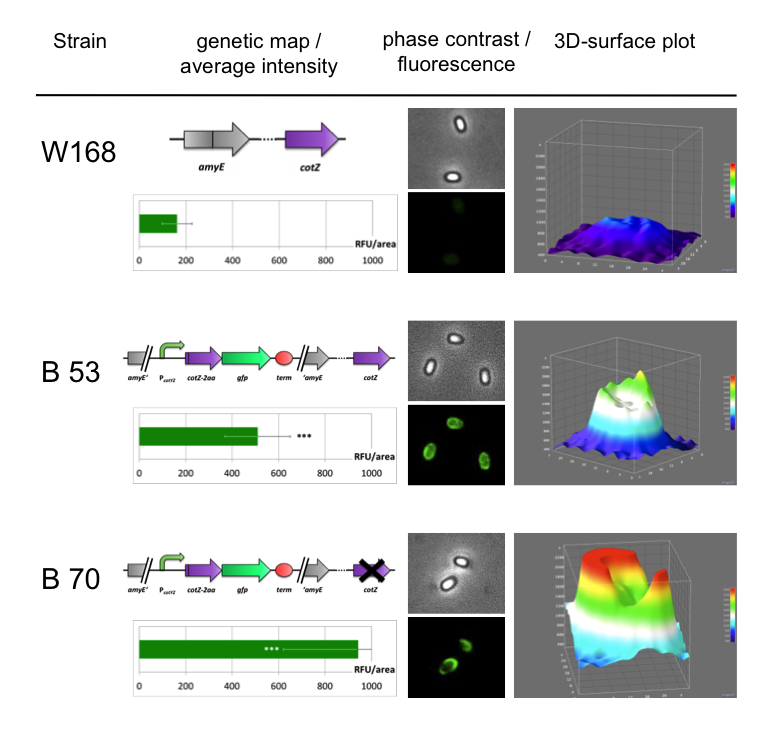
| + | <font color="#000000"; size="2">Result of fluorescence evaluation of the three strains: W168, B53 and B70.</font> | ||
| + | |} | ||
| + | |} | ||
| - | |||
| + | ===Erase<sup>*</sup> germination ability! Knockout strains=== | ||
| + | <br> | ||
{| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
{|align:center | {|align:center | ||
| - | |[[File: | + | |[[File:Germination_RatesIIii_ii.jpg|600px|center]] |
|- | |- | ||
| style="width: 80%;background-color: #EBFCE4;" | | | style="width: 80%;background-color: #EBFCE4;" | | ||
{| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
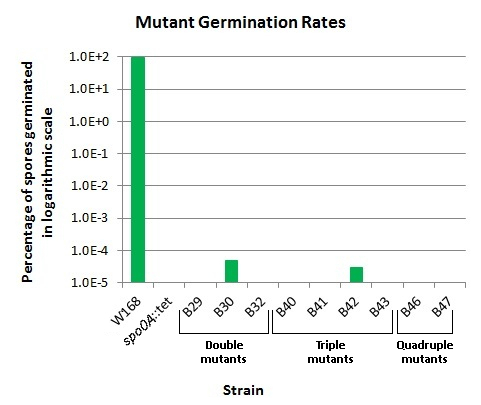
| - | <font color="#000000"; size="2">'''The germination rates of ''B. subtilis'' germination mutants.''' All germination rates are relative to wild type 168 ( | + | <font color="#000000"; size="2">'''The germination rates of ''B. subtilis'' germination mutants.''' All germination rates are relative to wild type 168 (W168), our positive control. W168 showed near 100% germination rates. The mutant ''spo0A''::tet is unable to form spores, and should therefore show no germination (our negative control). Our results were consistent with this expectation. Mutants used were: <br /> |
| - | ''' | + | '''W168''': wild-type 168 <br /> |
| - | '''''spo0A''::tet''': | + | '''''spo0A''::tet''': a sporulation-deficient strain <br /> |
'''B29''': ''cwlD''::kan, ''sleB''::mls <br /> | '''B29''': ''cwlD''::kan, ''sleB''::mls <br /> | ||
'''B30''': ''gerD''::cm, ''sleB''::mls <br /> | '''B30''': ''gerD''::cm, ''sleB''::mls <br /> | ||
| Line 126: | Line 139: | ||
'''B43''': ''gerD''::cm, ''sleB''::mls, ''cwlJ''::spec <br /> | '''B43''': ''gerD''::cm, ''sleB''::mls, ''cwlJ''::spec <br /> | ||
'''B46''': ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cm, ''sleB''::mls <br /> | '''B46''': ''cwlD''::kan, ''cwlJ''::spec, ''gerD''::cm, ''sleB''::mls <br /> | ||
| - | '''B47''': ''gerD''::cm, ''sleB''::mls, ''cwlJ''::spec, ''cwlD''::kan</font> | + | '''B47''': ''gerD''::cm, ''sleB''::mls, ''cwlJ''::spec, ''cwlD''::kan<br></font> |
|} | |} | ||
|} | |} | ||
|} | |} | ||
| + | <p align="justify">*No germinating spores detected in 4.6x10<sup>9</sup> spores. For a full explanation of these great results, see [https://2012.igem.org/Team:LMU-Munich/Data/Knockout All results].</p> | ||
| - | + | ===Inverter works!=== | |
| - | [[File:LMU Inverter graph.png|600px]] | + | <br> |
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:LMU Inverter graph.png|600px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
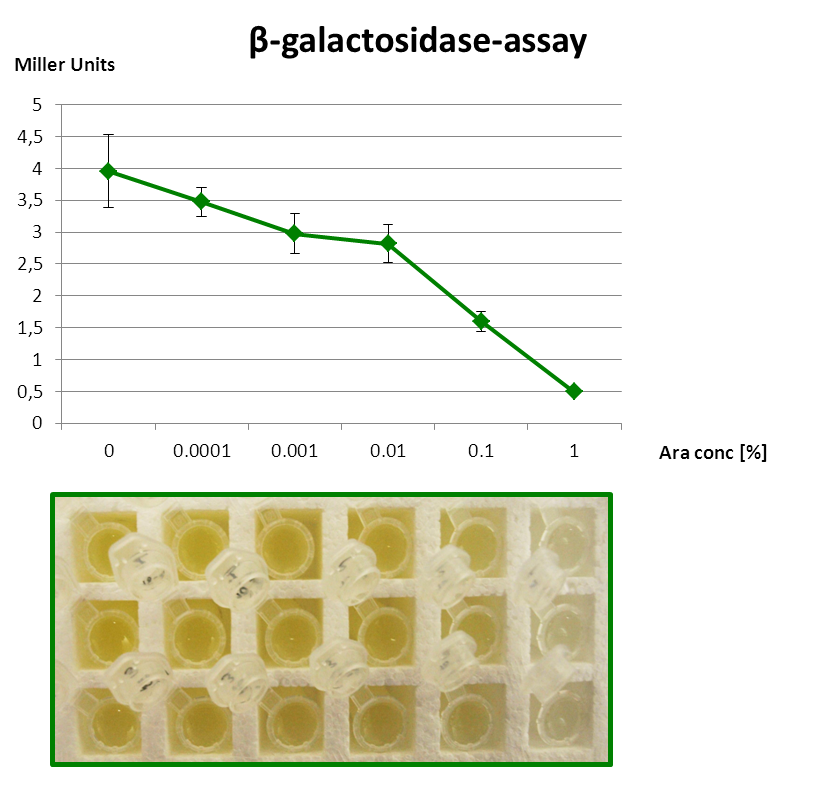
| + | <font color="#000000"; size="2"><p align="justify"> β-Galactosidase assay of Inverter with ''lacZα'' as reporter. The higher the arabinose concentration, the higher the translational repression of ''uof<sub>CGU</sub>-lacZα'' </p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
For detailed results visit our [[Team:LMU-Munich/Data|data page]] and for all background info our [[Team:LMU-Munich/Project|project page]]. | For detailed results visit our [[Team:LMU-Munich/Data|data page]] and for all background info our [[Team:LMU-Munich/Project|project page]]. | ||
| + | |||
| + | |||
| Line 153: | Line 182: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
| - | |||
Latest revision as of 22:13, 26 September 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Here you will find the main results of our work.
Seven novel vectors were constructed and four already proven to work in B. subtilis.
| Name
BioBrick | E. coli res. | B. subt. res. | Insertion locus | Description | Cloning status | Works? |
|---|---|---|---|---|---|---|
| pSBBs1C [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823023 (BBa_K823023)] | Amp | Cm | amyE | empty | ||
| pSBBs4S [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823022 (BBa_K823022)] | Amp | Spec | thrC | empty | ||
| pSBBs2E [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823027 (BBa_K823027)] | Amp | MLS | lacA | empty | in work | not tested |
| pSBBs1C-lacZ [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823021 (BBa_K823021) ] | Amp | Cm | amyE | lacZ reporter | ||
| pSBBs3C-luxABCDE [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 (BBa_K823025)] | Amp | Cm | sacA | luxABCDE reporter | ||
| pSBBs4S-PXyl [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823024 (BBa_K823024)] | Amp | Spec | thrC | Xylose-promoter | refuses transformation | |
| pSBBs0K-Pspac [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 (BBa_K823026)] | Amp | Kan | replicative | IPTG-promoter | unexpected | |
| Sporovector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 (BBa_K823054)] | Amp | Spec | thrC | to make Sporobeads | not tested |
15 Promoters evaluated in B. subtilis and added to the registry!
This section gives an overview on the strength of all evaluated promoters, which span a large range of activities. For more details and informations of the experiments see the Data page of the promoters. Note that Pveg was not evaluated with luminescence measurements and this bar is just projected from the results of the β-galactosidase assay.
|
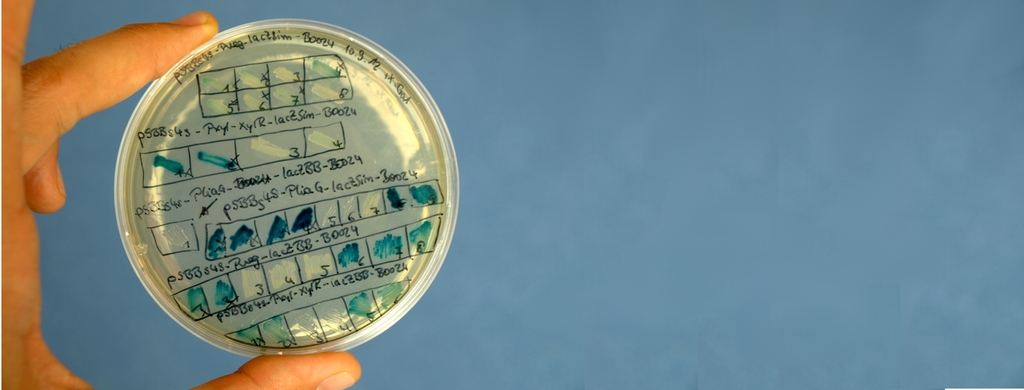
Sporobeads - what protein do you want to display?
|
| |
|
Erase* germination ability! Knockout strains
|
*No germinating spores detected in 4.6x109 spores. For a full explanation of these great results, see All results.
Inverter works!
|
For detailed results visit our data page and for all background info our project page.
 "
"