Team:LMU-Munich/Data/integration
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| (4 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
| - | [[File:Bacilluss_Intro.png|100px|right]] | + | [[File:Bacilluss_Intro.png|100px|right|link=]] |
| Line 10: | Line 10: | ||
===Transformation of ''B. subtilis''=== | ===Transformation of ''B. subtilis''=== | ||
<br> | <br> | ||
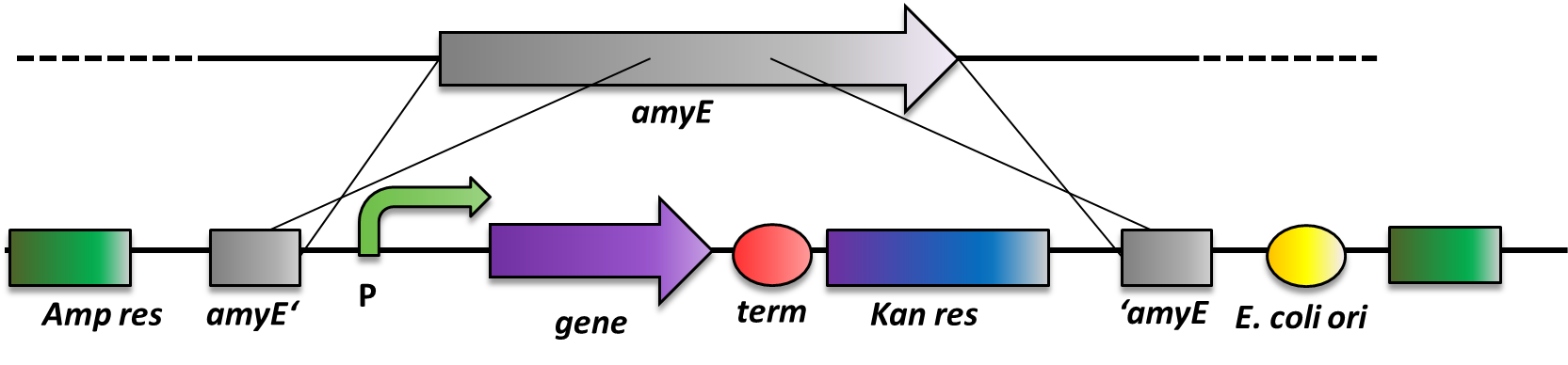
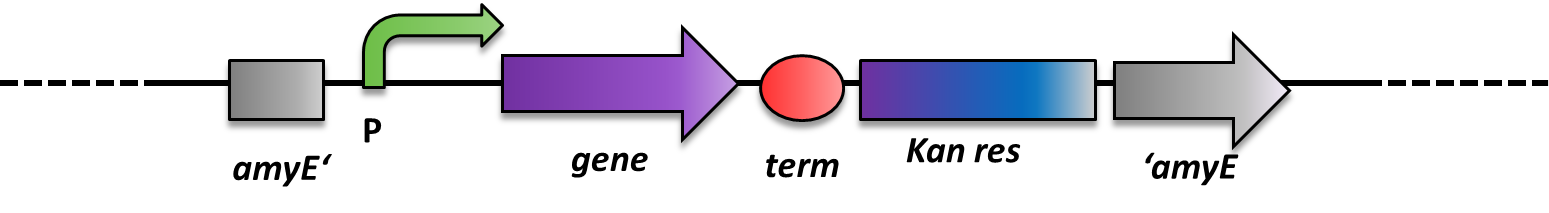
| - | <p align="justify">As ''B. subtilis'' and ''E. coli'' are model organisms, they have established genetics. The advantage of ''B. subtilis'' is that it is naturally competent | + | <p align="justify">As ''B. subtilis'' and ''E. coli'' are model organisms, they have established genetics. The advantage of ''B. subtilis'' is that it is naturally competent, so it is very easy to conduct genetic manipulations. It can replicate plasmids as ''E. coli'' does, but there is a much more elegant way of bringing in exogenous DNA fragments. When flanked by regions homologous to the ''B. subtilis'' genome, it will be integrated at high efficiency via homologous recombination at this locus and subsequently be replicated with the chromosome (Fig. 2). This leads to stable, single-copy genomic alterations, thereby avoiding, copy-number artifacts occurring with replicative plasmids. This different way of genetic manipulations requires the use of integrative vectors as provided by our [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''Bacillus'''''<b>B</b>io<b>B</b>rick<b>B</b>ox]. For this reason, ''B. subtilis'' is an ideal genetic platform for Synthetic Biology. But so far, very few iGEM teams have worked with this model organism due to the lack of suitable BioBrick-compatible genetic tools.</p> |
<br> | <br> | ||
| Line 37: | Line 37: | ||
| - | [[File:LMU Arrow purple BACK.png| | + | {| width="100%" cellpadding="20" |
| + | |[[File:LMU Arrow purple BACK.png|right|80px|link=Team:LMU-Munich/Bacillus_Introduction]] | ||
| + | |[[File:LMU Arrow purple NEXT.png|left|80px|link=Team:LMU-Munich/Data/differentiation]] | ||
| + | |} | ||
| + | |||
Latest revision as of 10:47, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]



Transformation of B. subtilis
As B. subtilis and E. coli are model organisms, they have established genetics. The advantage of B. subtilis is that it is naturally competent, so it is very easy to conduct genetic manipulations. It can replicate plasmids as E. coli does, but there is a much more elegant way of bringing in exogenous DNA fragments. When flanked by regions homologous to the B. subtilis genome, it will be integrated at high efficiency via homologous recombination at this locus and subsequently be replicated with the chromosome (Fig. 2). This leads to stable, single-copy genomic alterations, thereby avoiding, copy-number artifacts occurring with replicative plasmids. This different way of genetic manipulations requires the use of integrative vectors as provided by our BacillusBioBrickBox. For this reason, B. subtilis is an ideal genetic platform for Synthetic Biology. But so far, very few iGEM teams have worked with this model organism due to the lack of suitable BioBrick-compatible genetic tools.
|
 "
"