Team:LMU-Munich/Data/Vectors
From 2012.igem.org
| Line 114: | Line 114: | ||
<div class="box"> | <div class="box"> | ||
| - | ===pSB<sub>Bs</sub>1C-<i> | + | ===pSB<sub>Bs</sub>1C-<i>lac</i>Z=== |
<p align="justify"> | <p align="justify"> | ||
<html> <a> <img src="https://static.igem.org/mediawiki/2012/0/04/LMU-Munich-PSBBs1C-lacZ.png" height=130"/></a></html> | <html> <a> <img src="https://static.igem.org/mediawiki/2012/0/04/LMU-Munich-PSBBs1C-lacZ.png" height=130"/></a></html> | ||
| + | pSB<sub>Bs</sub>1C-<i>lac</i>Z is a reporter vector which includes a ribosome binding site, a β-galactosidase (''lac''Z) and a terminator downstream of the multiple cloning site. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. This vector integrates into the ''amy''E locus which is tested by the starch test. | ||
| + | This vector was used for several promoter evaluations. | ||
</p> | </p> | ||
</div> | </div> | ||
Revision as of 15:40, 21 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Bacillus subtilis vectors
For the detailed protocols, please see our protocol section.
Overview
| Name BioBrick | E. coli res. | B. subt. res. | Insertion locus | Description | Cloning status | Works? |
|---|---|---|---|---|---|---|
| pSBBs1C (BBa_K823023) | Amp | Cm | amyE | empty | ||
| pSBBs4S (BBa_K823022) | Amp | Spec | thrC | empty | ||
| pSBBs2E (BBa_K823027) | Amp | MLS | lacA | empty | in work | not tested |
| pSBBs1C-lacZ (BBa_K823021) | Amp | Cm | amyE | with lacZ reporter gene | ||
| pSBBs3C-luxABCDE (BBa_K823025) | Amp | Cm | sacA | with luxABCDE reporter cassette | ||
| pSBBs4S-PXyl (BBa_K823024) | Amp | Spec | thrC | with Xylose-inducible promoter | unexpected | |
| pSBBs0K-Pspac (BBa_K823026) | Amp | Kan | replicative | with IPTG inducible promoter | refuses transformation |
pSBBs1C
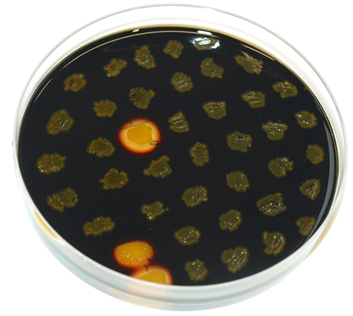
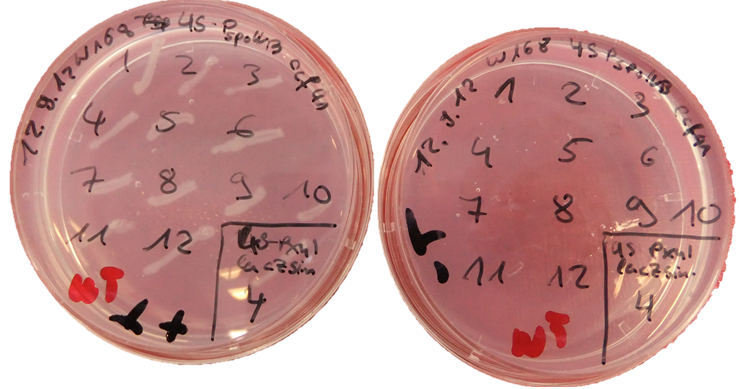
pSBBs1C is one of our empty vectors. It integrates into the amyE locus of Bacillus subtilis and carries a Chloramphenicol resistance as well as an Ampicillin resistance for E. coli. Inbetween the two recombination sites, there is the constitutively expressed Cm resistance and the multiple cloning site which contains RFP to simplify the insertion of your BioBrick.The plasmid is then transformed into B. subtilis and checked via the resistance for "integration at all" and via a starch test for "integration into the right locus". Please note that the colonies without a bright surounding are correct.
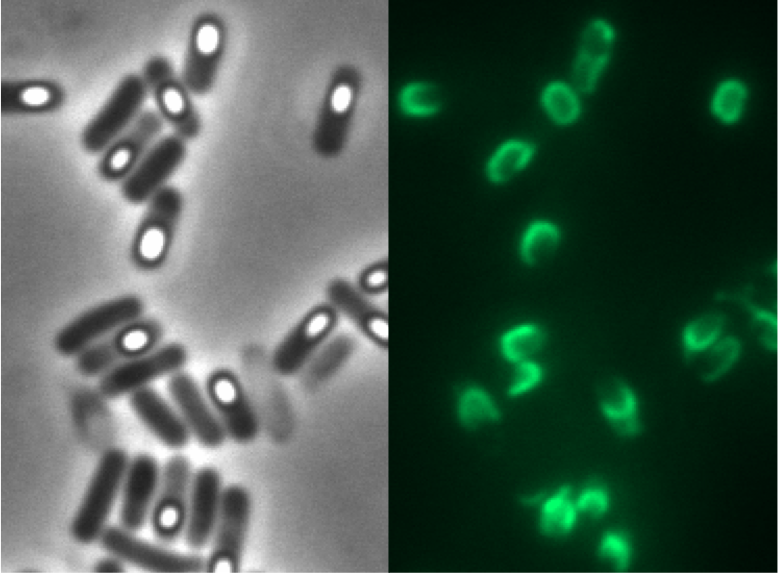
We tested this vector with various inserts, for example the final construct of our Sporobeads with GFP. The starch test shows that the insertion in in the amyE locus and fluorescence microscopy reveals the functionality of the construct.
pSBBs4S
pSBBs4S also is an empty vector with only the multiple cloning site and the spectinomycine resistence gene in between the homologous regions. This vector integrates into the thrC locus of B. subtilis.
We tested the integration of this plasmid with the threonine test and a construct from the Suicideswitch as insert.
pSBBs2E
 pSBBs2E is our third empty vector, aiming to provide a set of three empty vectors with three different integration loci and three different resistances for full flexibility in their combinations.
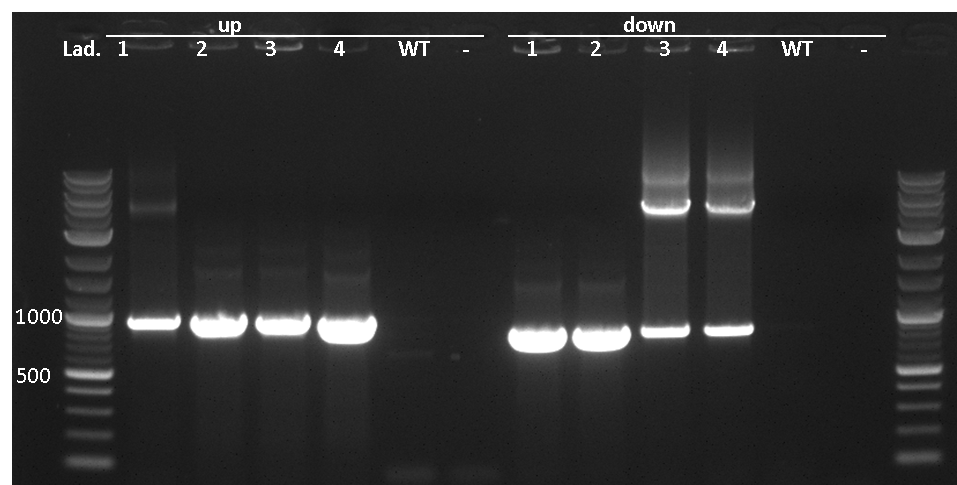
This vector is still in work. By now, only the multiple cloning site is missing. Of course, it is not tested yet.
pSBBs2E is our third empty vector, aiming to provide a set of three empty vectors with three different integration loci and three different resistances for full flexibility in their combinations.
This vector is still in work. By now, only the multiple cloning site is missing. Of course, it is not tested yet.
pSBBs1C-lacZ
 pSBBs1C-lacZ is a reporter vector which includes a ribosome binding site, a β-galactosidase (lacZ) and a terminator downstream of the multiple cloning site. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. This vector integrates into the amyE locus which is tested by the starch test.
This vector was used for several promoter evaluations.
pSBBs1C-lacZ is a reporter vector which includes a ribosome binding site, a β-galactosidase (lacZ) and a terminator downstream of the multiple cloning site. Also, a chloramphenicol resistance is in between the two flanking homolgy regions. This vector integrates into the amyE locus which is tested by the starch test.
This vector was used for several promoter evaluations.
 "
"