Team:LMU-Munich/Data/Anderson
From 2012.igem.org
| (11 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
| - | + | ==Anderson Promoters== | |
| - | + | <br> | |
===Luminescence measurements=== | ===Luminescence measurements=== | ||
| - | |||
<p align="justify"> | <p align="justify"> | ||
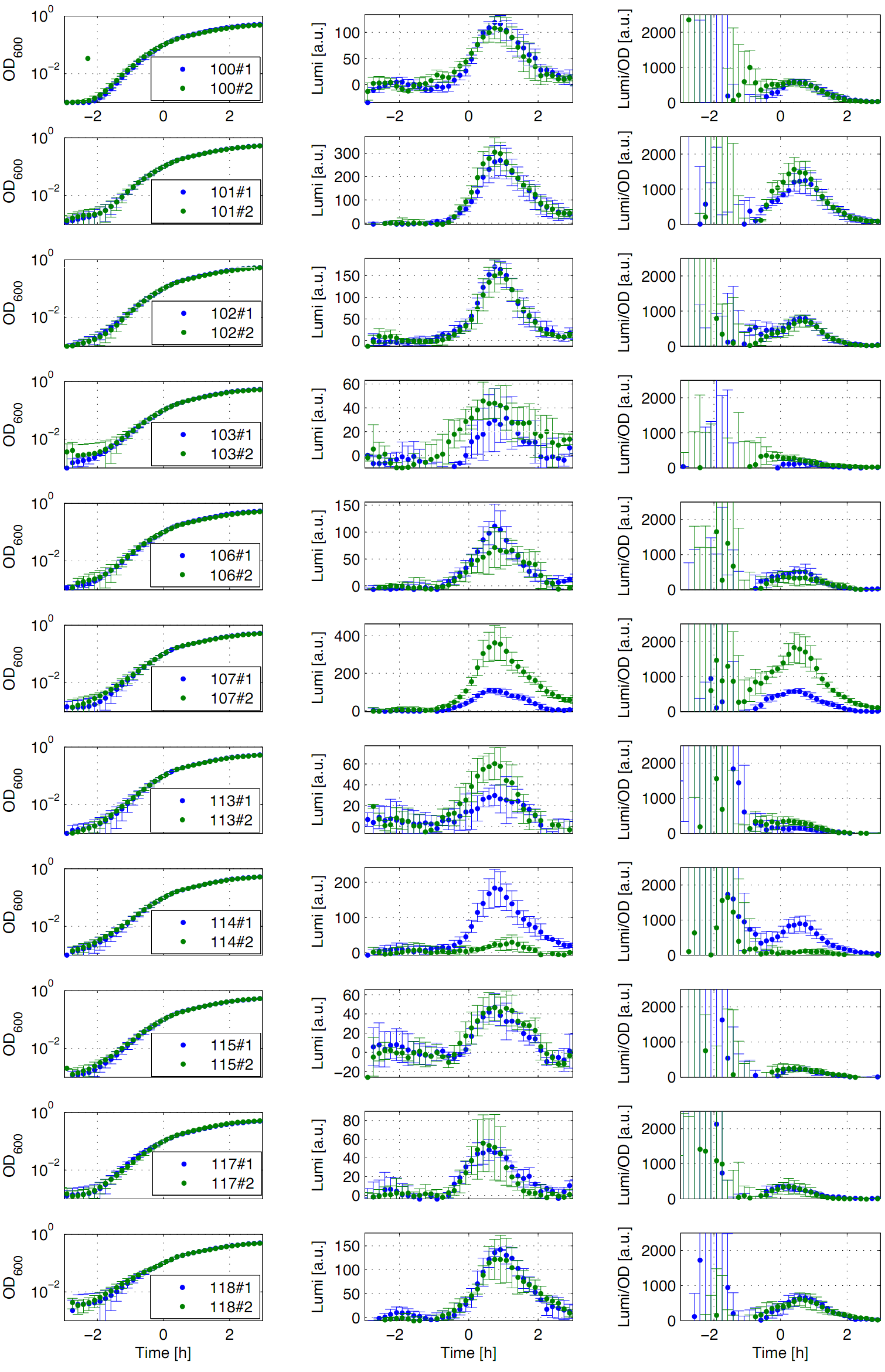
| - | Eleven of the nineteen promoters of the [http://partsregistry.org/Part:BBa_J23100 '''Anderson collection'''] (J23100, J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) were evaluated. | + | Eleven of the nineteen promoters of the [http://partsregistry.org/Part:BBa_J23100 '''Anderson collection'''] (J23100, J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) were evaluated. For that purpose, we used the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' from the BioBrickBox containing the ''lux'' operon [[File:Lux operon.png|100px]] as a reporter for promoter activity. The promoter activity leads to the expression of the ''lux'' operon and to the production of the enzyme luciferase. The bioluminescence, which is produced by the luciferase, can be measured with the microtiter plate reader ''Synergy2'' ([http://www.biotek.com/ BioTek]) ('''Fig.1''').</p> |
{| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| Line 23: | Line 22: | ||
{| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
| - | <font color="#000000"; size="2"><p align="justify"> '''Fig. 1: Luminescence measurement of Anderson promoters in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'''''. OD<sub>600</sub> (right), Lumi (middle) and Lumi per OD<sub>''600''</sub> (left) depending on the time (h) are shown for two different clones (green/blue). Data | + | <font color="#000000"; size="2"><p align="justify"> '''Fig. 1: Luminescence measurement of Anderson promoters in the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'''''. OD<sub>600</sub> (right), Lumi (middle) and Lumi per OD<sub>''600''</sub> (left) depending on the time (h) are shown for two different clones (green/blue). Data derived from three independent experiments, graph shows the mean with standard deviation. Curves were fitted over each other (t=0, OD<sub>600</sub>=0,3) and smoothed by taking average of three neighboring values.</p></font> |
|} | |} | ||
|} | |} | ||
|} | |} | ||
<p align="justify"> | <p align="justify"> | ||
| - | All clones show a | + | All clones show a typical growth behaviour. The activity of the promoters increases during transition from log to stationary phase. The maximal promoter activity (t=1h) reaches 200 to 1500 Lumi/OD<sub>600</sub> for promoters J23115 and J23101, respectively. Afterwards, the activities drop to the initial levels (t=2h). The deviations of luminescence values (Lumi/OD<sub>600</sub>) in the beginning of the curves are an experimental artifact due to the small OD<sub>600</sub> values. One clone of J23107 and J23114 showed significantly lower promoter activities. Therefore, additional clones need to be measured. In comparison to all the other evaluated ''Bacillus'' promoters, the Anderson promoters showed a rather low acitivity in ''B. subtilis''.</p> |
<br> | <br> | ||
<br> | <br> | ||
| + | ===β-galactosidase assays=== | ||
<br> | <br> | ||
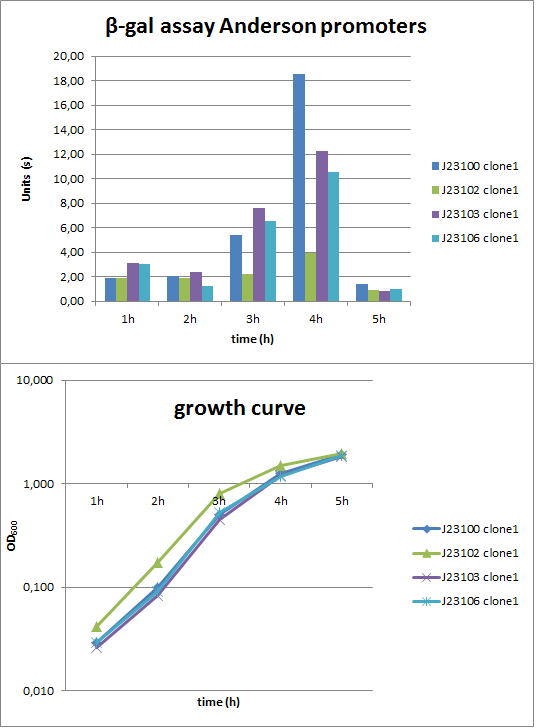
| - | + | <p align="justify">To evaluate the activity not only with the ''lux'' reporter, four promoters of the Anderson collection were cloned into the reporter vector pSB<sub>''Bs''</sub>1C-''lacZ'' [[File:LacZ.png|50px]] to perform β-galactosidase assays ('''Fig. 2'''). The results were then compared to the data of the luminescence measurements. | |
| - | + | ||
| - | + | ||
| - | <p align="justify">To evaluate the activity not only with the ''lux'' reporter | + | |
</p> | </p> | ||
| Line 47: | Line 44: | ||
{| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
| - | <font color="#000000"; size="2"><p align="justify"> '''Fig. 2:''' β-galactosidase assay (up) and growth curve (down) of the Anderson promoters J23100, J23102, J23103, J23106 in ''B. subtilis''. Results derive from one experiment and are only shown for one Bacillus clone. </p></font> | + | <font color="#000000"; size="2"><p align="justify"> '''Fig. 2:''' β-galactosidase assay (up) and growth curve (down) of the Anderson promoters J23100, J23102, J23103, J23106 in ''B. subtilis''. Results derive from one experiment and are only shown for one ''Bacillus'' clone. </p></font> |
|} | |} | ||
|} | |} | ||
|} | |} | ||
<br> | <br> | ||
| + | <p align="justify">Verification of the promoter activity by β-galactosidase assays revealed that the Anderson promoters do not seem to be as weak as measured by luminescence (Fig. 1). For a direct comparison, we need to include a constitutive promoter, e.g. P<sub>''liaG''</sub>, and repeat the same experiment. But we think the luminescence measurements are more reliable, because they were repeated three times with two independent clones.</p> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 57: | Line 55: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
[[File:Arrow purple Data.png|center|100px|link=Team:LMU-Munich/Data]] | [[File:Arrow purple Data.png|center|100px|link=Team:LMU-Munich/Data]] | ||
<!-- Include the next line at the end of every page --> | <!-- Include the next line at the end of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 01:52, 27 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Anderson Promoters
Luminescence measurements
Eleven of the nineteen promoters of the Anderson collection (J23100, J23101, J23102, J23103, J23106, J23107, J23113, J23114, J23115, J23117, J23118) were evaluated. For that purpose, we used the reporter vector pSBBs3C-luxABCDE from the BioBrickBox containing the lux operon ![]() as a reporter for promoter activity. The promoter activity leads to the expression of the lux operon and to the production of the enzyme luciferase. The bioluminescence, which is produced by the luciferase, can be measured with the microtiter plate reader Synergy2 (BioTek) (Fig.1).
as a reporter for promoter activity. The promoter activity leads to the expression of the lux operon and to the production of the enzyme luciferase. The bioluminescence, which is produced by the luciferase, can be measured with the microtiter plate reader Synergy2 (BioTek) (Fig.1).
All clones show a typical growth behaviour. The activity of the promoters increases during transition from log to stationary phase. The maximal promoter activity (t=1h) reaches 200 to 1500 Lumi/OD600 for promoters J23115 and J23101, respectively. Afterwards, the activities drop to the initial levels (t=2h). The deviations of luminescence values (Lumi/OD600) in the beginning of the curves are an experimental artifact due to the small OD600 values. One clone of J23107 and J23114 showed significantly lower promoter activities. Therefore, additional clones need to be measured. In comparison to all the other evaluated Bacillus promoters, the Anderson promoters showed a rather low acitivity in B. subtilis.
β-galactosidase assays
To evaluate the activity not only with the lux reporter, four promoters of the Anderson collection were cloned into the reporter vector pSBBs1C-lacZ ![]() to perform β-galactosidase assays (Fig. 2). The results were then compared to the data of the luminescence measurements.
to perform β-galactosidase assays (Fig. 2). The results were then compared to the data of the luminescence measurements.
|
Verification of the promoter activity by β-galactosidase assays revealed that the Anderson promoters do not seem to be as weak as measured by luminescence (Fig. 1). For a direct comparison, we need to include a constitutive promoter, e.g. PliaG, and repeat the same experiment. But we think the luminescence measurements are more reliable, because they were repeated three times with two independent clones.
 "
"