|
|
| (48 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | <!-- Include the next line at the beginning of every page --> | | <!-- Include the next line at the beginning of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} | | {{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} |
| - | [[File:Bacillus_introduction_banner.jpg|620px|link=]] | + | [[File:Bacillus introduction banner.resized WORDS.JPG|620px|link=]] |
| - | [[File:Bacilluss Intro.png|100px|right]]
| + | |
| - | <p></p>
| + | |
| | | | |
| | + | |
| | + | [[File:Bacilluss_Intro.png|100px|right|link=]] |
| | | | |
| | | | |
| | | | |
| | ==''Bacillus subtilis'' - a new chassis for iGEM== | | ==''Bacillus subtilis'' - a new chassis for iGEM== |
| - | | + | <br> |
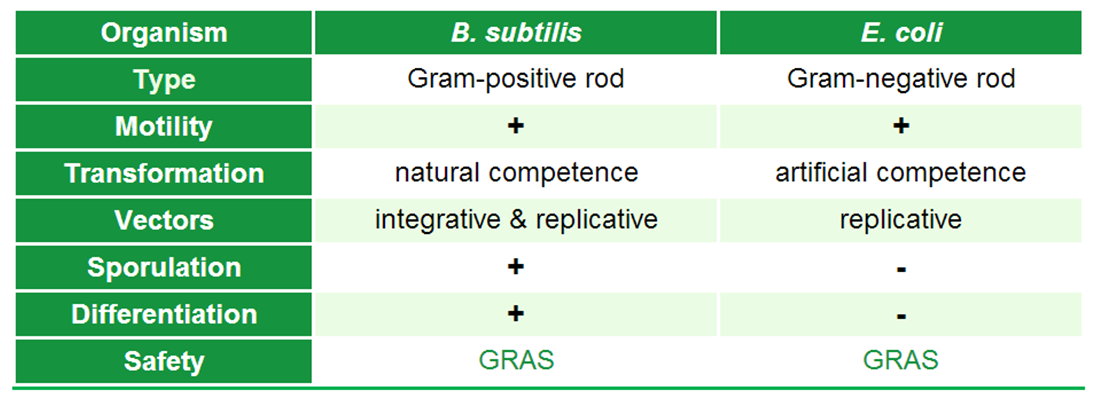
| - | <p align="justify">We chose to work with ''Bacillus subtilis'' to set new horizons in the ''Escherichia coli''-dominated world of iGEM by offering a set of tool for this gram-positive model organism to the . To introduce ''B. subtilis'', we first want to highlight some important aspects of this organism, which are listed in the table below.</p> | + | <div class="box"> |
| - | | + | ====Introduction==== |
| - | | + | {| width="100%" style="text-align:center;"| |
| - | {| class="colored" width="100%" style="text-align:center;" | + | |<p align="justify">Introduction to ''B. subtilis'' and background information.</p> |
| - | |- | + | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Data/introintro]] |
| - | !Organism
| + | |
| - | !''B. subtilis''
| + | |
| - | !''E. coli''
| + | |
| - | |- | + | |
| - | !<font color="#EBFCE4">Type</font>
| + | |
| - | |gram-positive rod
| + | |
| - | |gram-negative rod
| + | |
| - | |-
| + | |
| - | !Motility
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |- | + | |
| - | !Transformation
| + | |
| - | |natural competence | + | |
| - | |artificial competence | + | |
| - | |- | + | |
| - | !Vectors
| + | |
| - | |integrative & replicative
| + | |
| - | |replicative
| + | |
| - | |-
| + | |
| - | !Sporulation
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |<font size=4>'''-'''
| + | |
| - | |-
| + | |
| - | !Differentiation
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |<font size=4>'''-'''
| + | |
| - | |-
| + | |
| - | !Safety
| + | |
| - | |[http://www.fda.gov/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/default.htm GRAS]
| + | |
| - | |[http://www.fda.gov/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/default.htm GRAS]
| + | |
| | |- | | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/introintro]] |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | <br>
| |
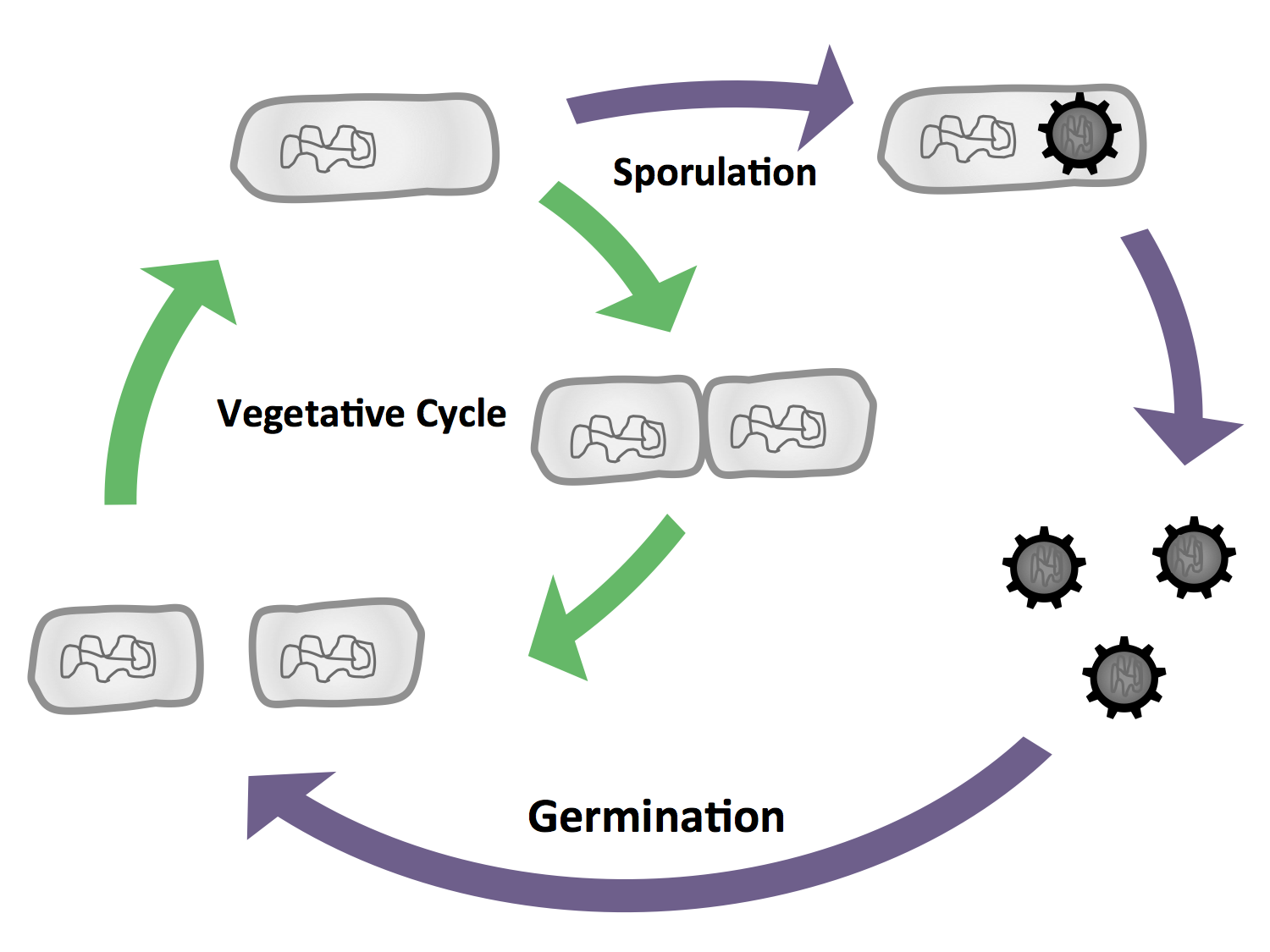
| - | <p align="justify"> In general, bacteria can be divided into two major groups that differ primarily differ in their cell envelope: gram-positive and gram-negative. ''E. coli'' is the model organism for the gram-negative bacteria. A model organism for the gram-positive bacteria is ''B. subtilis'', our favourite pet. The natural habitat of ''B. subtilis'' is the soil, so it is forced to adapt to numerous environmental changes. Accordingly, ''B. subtilis'' is characterized by quick and cunning reflexes and a complex life style. There are many differentiations and survival strategies that ''B. subtilis'' can engage (Fig. 1): Due to its natural competence, it can take up exogenous DNA and integrate it into its genome. To be flexibel to the environment and move towards nutrients or avoid toxic compounds, it is motile with the aid of its peritrichous flagella. If starved some cells even become cannibles that fees on their siblings. If the conditions get too bad for survival, ''B. subtilis'' can form endospores. These are very dormant and highly stable stages that are resistent against e.g. desiccation, UV light, heat and pressure. Once these spores encounter better conditions they are able to germinate again. See this [section] for Details.</p>
| |
| | | | |
| | + | There are two major differences between ''B. subtilis'' and ''E. coli'' that are of interest to us: |
| | | | |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| + | <div class="box"> |
| - | | style="width: 70%;background-color: #EBFCE4;" | | + | ====Transformation of ''B. subtilis''==== |
| - | {|
| + | {| width="100%" style="text-align:center;"| |
| - | |[[File:LMU Bacillus life.png|center|450px]] | + | |<p align="justify">Cloning strategy for the work of '' B. subtilis''.</p> |
| | + | |[[File:Integration.png|right|150px|link=Team:LMU-Munich/Data/integration]] |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/integration]] |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 1: Schematic representation of the distinct cell types that differentiate in the communities of ''Bacillus subtilis''.<br> ([http://www.ncbi.nlm.nih.gov/pubmed/20030732 Lopez & Kolter, 2010])</p></font>
| + | |
| - | |}
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | | + | <div class="box"> |
| - | <p align="justify"> | + | ====Differentiation==== |
| - | There are two major differences between ''B. subtilis'' and ''E. coli'' that are of interest to us:
| + | {| width="100%" style="text-align:center;"| |
| - | <br>
| + | |<p align="justify">The life cycle of ''B. subtilis'' and the different cell stages.</p> |
| - | <br>
| + | |[[File:Figures Bacillus Intro fig1.png|right|150px|link=Team:LMU-Munich/Data/differentiation]] |
| - | '''1) Transformation of ''B. subtilis'''''
| + | |
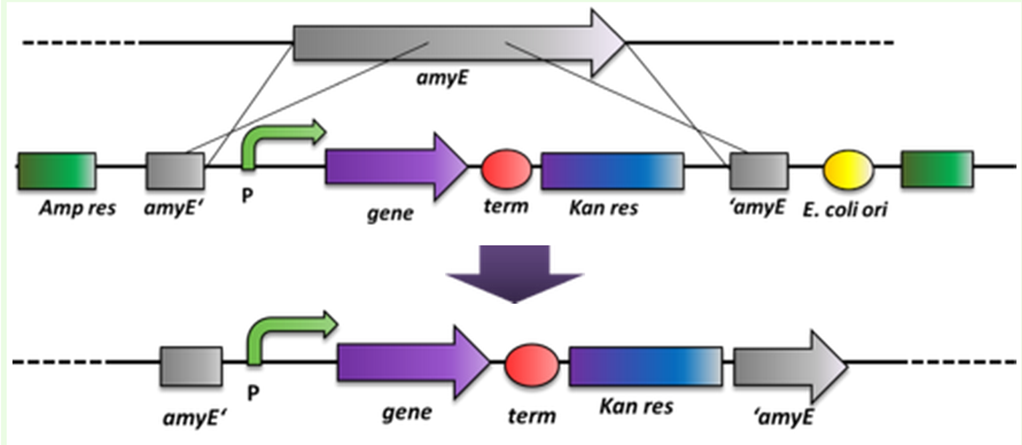
| - | <br>As ''B. subtilis'' and ''E. coli'' are model organisms, they have established genetics. The advantage of ''B. subtilis'' is that it is naturally competent. So it is not complicated to conduct genetical manipulations. It can replicate exogenous DNA via an origin of replication on a plasmid as ''E. coli'' does, but there is a much more elegant way of bringing in exogenous DNA stretches. When flanked by homologous regions to the bacterial genome, it will be integrated at high efficiency via homologous recombination at this locus and furthermore be replicated with the genome. This has the advantage that if comparing different variables, not only the enviroment is always the same, but also the copy number is from cell to cell and from strain to strain the same, which is not always the case for replicative plasmids. This integrative way of bringing in exogenous DNA was exploited by us when producing the BioBrick compatible ''Bacillus'' vectors. The comparision between these two ways of bringing in exogenous DNA is depicted in Fig. 2. For these reasons, in some cases ''B. subtilis'' can be the chassis of choice. Unfortunately, very few iGEM teams have worked with this model organism, and there is at this time no established BioBrick system to use ''B. subtilis'' as a chassis.</p>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |
| - | | style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | {|
| + | |
| - | {|
| + | |
| - | |[[File:LMU-Munich-insertion-Intro1.png|500px|center]]
| + | |
| - | |-
| + | |
| - | |[[File:LMU-Munich-integration-Arrow.png|70px|center]] | + | |
| - | |- | + | |
| - | |[[File:LMU-Munich-insertion-Intro2.png|500px|center]]
| + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/differentiation]] |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 2: Exogenous DNA is shown in red while the bacterial genome is black a) The propagation of exogenous DNA if brought in as replicative plasmid. The number of plasmids per cell can vary. b) The propagation of exogenous DNA if it is able to integrate into the genome via homologous recombination</p></font>
| + | |
| | |} | | |} |
| - | |}
| + | </div> |
| - | |}
| + | <br> |
| - | | + | |
| | <br> | | <br> |
| | <br> | | <br> |
| - | '''2) Differentiation'''
| |
| - | <p align="justify">''B. subtilis'' is able to differentiate into cells with different morphology and function (Fig. 3), the most severe form being the endospore which is produced under stress conditions. These spores are resistant towards environmental influences. But if they sense favour conditions they can germinate again. In our project, we will exploit the production of endospores. Because they are extremely stable, they are good vehicles for our fusion proteins with certain functions.
| |
| | <br> | | <br> |
| - | </p>
| |
| - |
| |
| - |
| |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {|
| |
| - | |[[File:Figures Bacillus Intro fig1.png|500px|center]]
| |
| - | |-
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 3: The vegetative cycle is very similiar to the one of ''E. coli.'' But if there is a stress like for example starvation, the cells enter sporulation, where they first undergo a polar cell division, followed by the formation of the endospore. If the enviromental conditions are suitable again, the spore will then germinate and reenter the vegetative cycle.</p></font>
| |
| - | |}
| |
| - | |}
| |
| - | |}
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| | <div class="box"> | | <div class="box"> |
| | ====Project Navigation==== | | ====Project Navigation==== |
| Line 126: |
Line 52: |
| | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] | | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] |
| | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] |
| - | |[[File:TerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] | + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] |
| | |- | | |- |
| | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] | | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] |
| Line 134: |
Line 60: |
| | |} | | |} |
| | </div> | | </div> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | | | |
| | {{:Team:LMU-Munich/Templates/Page Footer}} | | {{:Team:LMU-Munich/Templates/Page Footer}} |








 "
"