Team:LMU-Munich/Bacillus Introduction
From 2012.igem.org
(Difference between revisions)
| (137 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} | {{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} | ||
| + | [[File:Bacillus introduction banner.resized WORDS.JPG|620px|link=]] | ||
| - | |||
| + | [[File:Bacilluss_Intro.png|100px|right|link=]] | ||
| - | |||
| - | + | ==''Bacillus subtilis'' - a new chassis for iGEM== | |
| + | <br> | ||
| + | <div class="box"> | ||
| + | ====Introduction==== | ||
| + | {| width="100%" style="text-align:center;"| | ||
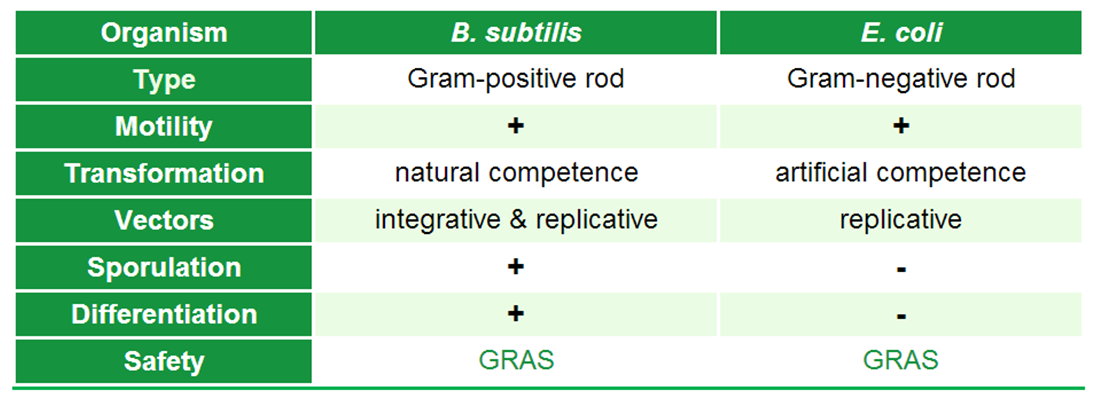
| + | |<p align="justify">Introduction to ''B. subtilis'' and background information.</p> | ||
| + | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Data/introintro]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/introintro]] | ||
| + | |} | ||
| + | </div> | ||
| - | + | There are two major differences between ''B. subtilis'' and ''E. coli'' that are of interest to us: | |
| + | <div class="box"> | ||
| + | ====Transformation of ''B. subtilis''==== | ||
| + | {| width="100%" style="text-align:center;"| | ||
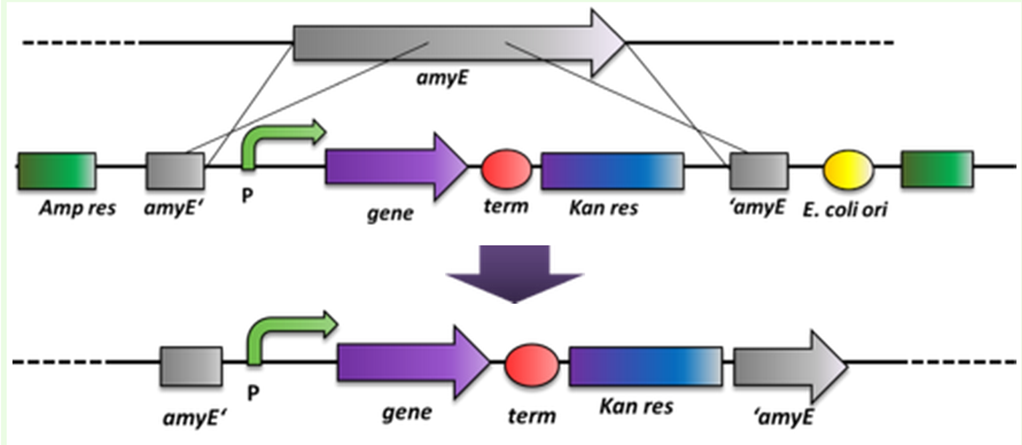
| + | |<p align="justify">Cloning strategy for the work of '' B. subtilis''.</p> | ||
| + | |[[File:Integration.png|right|150px|link=Team:LMU-Munich/Data/integration]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/integration]] | ||
| + | |} | ||
| + | </div> | ||
| - | + | <div class="box"> | |
| - | + | ====Differentiation==== | |
| - | + | {| width="100%" style="text-align:center;"| | |
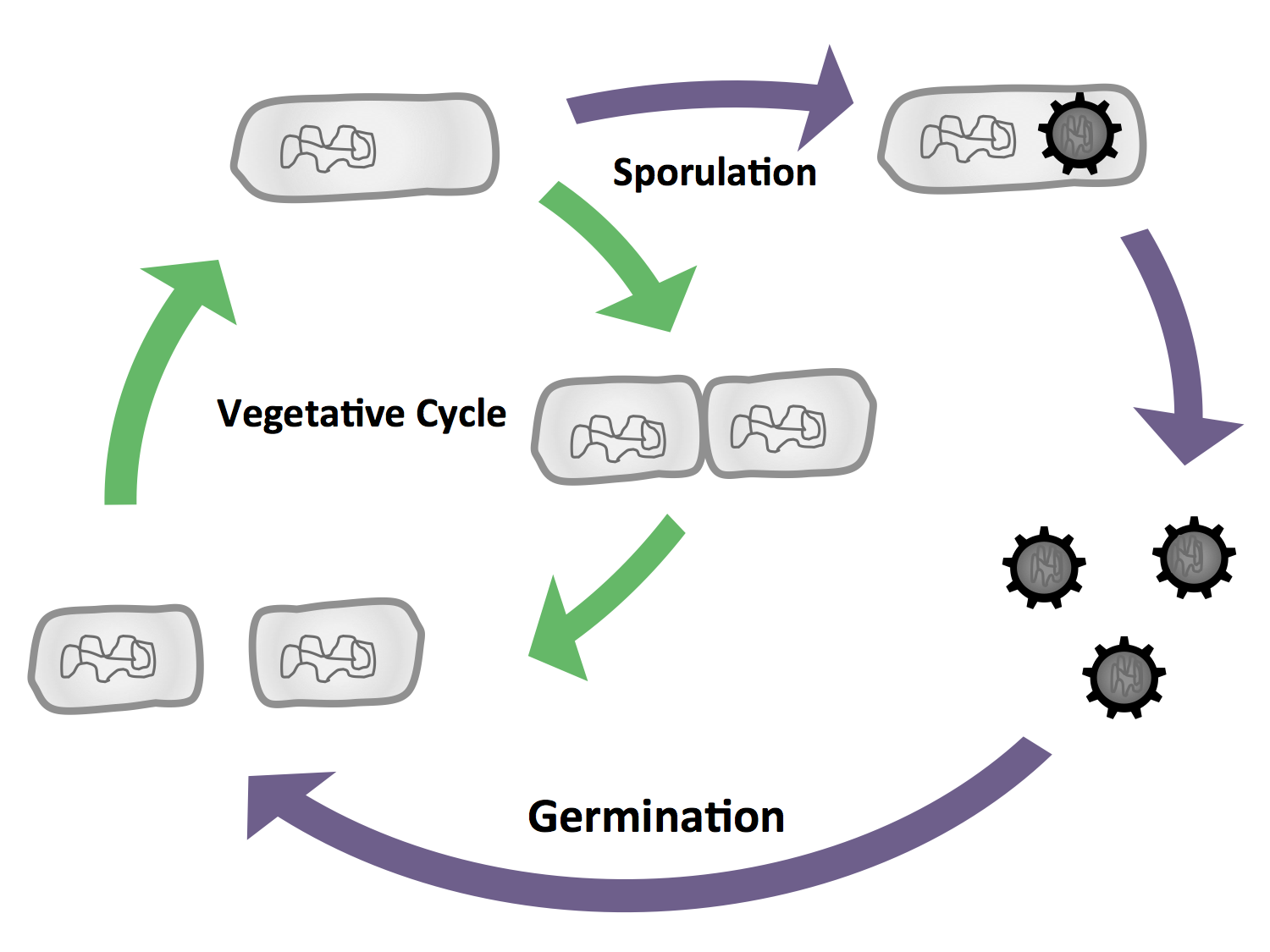
| - | + | |<p align="justify">The life cycle of ''B. subtilis'' and the different cell stages.</p> | |
| - | + | |[[File:Figures Bacillus Intro fig1.png|right|150px|link=Team:LMU-Munich/Data/differentiation]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |< | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/differentiation]] |
| - | | | + | |} |
| - | | | + | </div> |
| - | |GerminationSTOP | + | <br> |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div class="box"> | ||
| + | ====Project Navigation==== | ||
| + | {| width="100%" align="center" style="text-align:center;" | ||
| + | |[[File:Bacilluss_Intro.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] | ||
| + | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] | ||
| + | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] | ||
|- | |- | ||
| - | + | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] | |
| - | + | |[[Team:LMU-Munich/Bacillus_BioBricks|<font size="2" face="verdana">'''''Bacillus'''''<BR>'''B'''io'''B'''rick'''B'''ox</font>]] | |
| - | + | |[[Team:LMU-Munich/Spore_Coat_Proteins|<font size="2" face="verdana">'''Sporo'''beads</font>]] | |
| - | + | |[[Team:LMU-Munich/Germination_Stop|<font size="2" face="verdana">'''Germination'''<BR>STOP</font>]] | |
| - | + | |} | |
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 16:53, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]



Bacillus subtilis - a new chassis for iGEM
There are two major differences between B. subtilis and E. coli that are of interest to us:

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"