Team:LMU-Munich/Bacillus BioBricks/Reporters
From 2012.igem.org
| Line 53: | Line 53: | ||
Reference: [http://www.evrogen.com/products/mKate2/mKate2.shtml mkate2] | Reference: [http://www.evrogen.com/products/mKate2/mKate2.shtml mkate2] | ||
| + | |||
| + | |||
| + | *'''LacZ''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 BioBrick:BBa_K823019]) | ||
| + | |||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:LacZ plate.png|center|400px]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2"><p align="justify">''lacZ'' fused to the terminator B0014 under the control of P<sub>''spac''</sub> in the expression vector pSB<sub>Bs</sub>0K-P<sub>''spac''</sub>. P<sub>''spac''</sub> induced with IPTG</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | |||
| + | <p align="justify">This evaluated ''lacZ'' gene is derived from the ''Bacillus'' reporter vector pAC6. It is constructed in the Freiburg Standard ([http://partsregistry.org/Help:Assembly_standard_25 RFC 25]) for in-frame fusion proteins. It also includes a ribosome binding site optimized for ''Bacillus subtilis'' translation. This ''lacZ'' BioBrick was tested in the expression vector pSB<sub>''Bs''</sub>0K-P<sub>''spac''</sub>. This construct gave output, so the reporter gene is functional qualitatively. See [https://2012.igem.org/Team:LMU-Munich/Data/Vectors Data] in the vector evaluation section of pSB<sub>''Bs''</sub>0K-P<sub>''spac''</sub>. A more qualitatively measurement will follow soon.</p> | ||
| + | |||
| + | Reference for the vector: [http://www.ncbi.nlm.nih.gov/pubmed/11902727 pAC6] | ||
| + | |||
| + | |||
| + | |||
| + | *'''luc''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 BioBrick:BBa_K823028]) | ||
| + | <p align="justify"> | ||
| + | The <i>luc</i> gene encodes the monomeric firefly luciferase, which requires a special substrate to produce luminescence. We codon-optimized it for <i>Bacillus subtilis</i>, added a ribosome binding site and synthesized by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html GeneArt]. | ||
| + | </p> | ||
| + | <p align="justify"> | ||
| + | Firefly luciferase is by far the most commonly used bioluminescent reporter. This monomeric enzyme of 61kDa catalyzes a two-step oxidation reaction to yield light, usually in the green to yellow region, typically 550–570nm . The first step is activation of the luciferyl carboxylate by ATP to yield a reactive mixed anhydride. In the second step, this activated intermediate reacts with oxygen to create a transient dioxetane that breaks down to the oxidized products, oxyluciferin and CO<sub>2</sub>. Upon mixing with substrates, firefly luciferase produces an initial burst of light that decays over about 15 seconds to a low level of sustained luminescence. This kinetic profile reflects the slow release of the enzymatic product, thus limiting catalytic turnover after the initial reaction. | ||
| + | </p> | ||
| + | <p align="justify"> | ||
| + | The popularity of native firefly luciferase as a genetic reporter is due to the sensitivity and convenience of the enzyme assay and tight coupling of protein synthesis with enzyme activity. Firefly luciferase, which is encoded by the ''luc'' gene, is a monomer that does not require any post-translational modifications; it is available as a mature enzyme directly upon translation of its mRNA. Catalytic competence is attained immediately after release from the ribosome. Also, luciferase has a very short half-life in cells (approximately 3 hours). Combined, these properties make luciferase an extremely responsive reporter, far more so than other commonly used reporters. | ||
| + | </p> | ||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:Luciferin reaction mod.png|center|400px]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Reference:[http://www.promega.com/resources/product-guides-and-selectors/protocols-and-applications-guide/bioluminescent-reporters/ Promega]</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | <p align="justify">It was used in <i>B. subtilis</i> before ([http://www.ncbi.nlm.nih.gov/pubmed/21552330 Mirouze et al. 2011]). | ||
| + | The non-optimized version of this reporter gene was already successfully used in B. subtilis. While we have not found the time to evaluate this BioBrick we expect that it will work at least as good.</p> | ||
| + | |||
Revision as of 20:28, 24 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Bacillus Reporters 
We designed and codon-optimized a set of reporters that are commonly used in B. subtilis. All reporters have a modified iGEM Freiburg standard (RFC 25) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the B. subitlis optimized ribosome binding site.
Find out more about the design of our prefix with ribosome binding site.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
- GFP (BioBrick:BBa_K823039)
|
We took a gfp derivate of the Bacillus subtilis plasmid pGFPamy and added the BioBrick compatible pre- and suffix of the Freiburg standard (RFC 25). The functionality of this BioBrick verified by creat e.g. our Sporobeads.
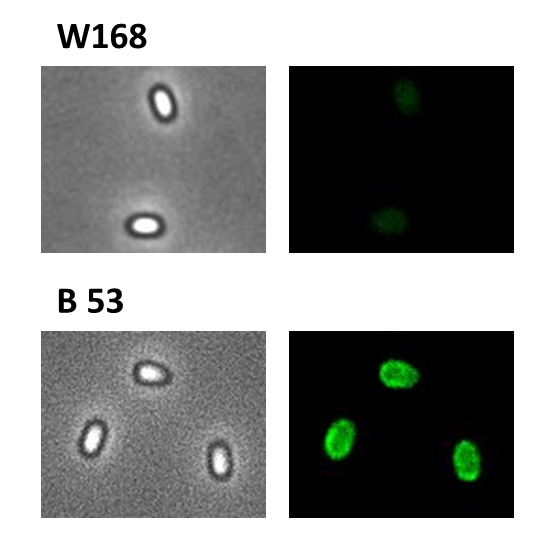
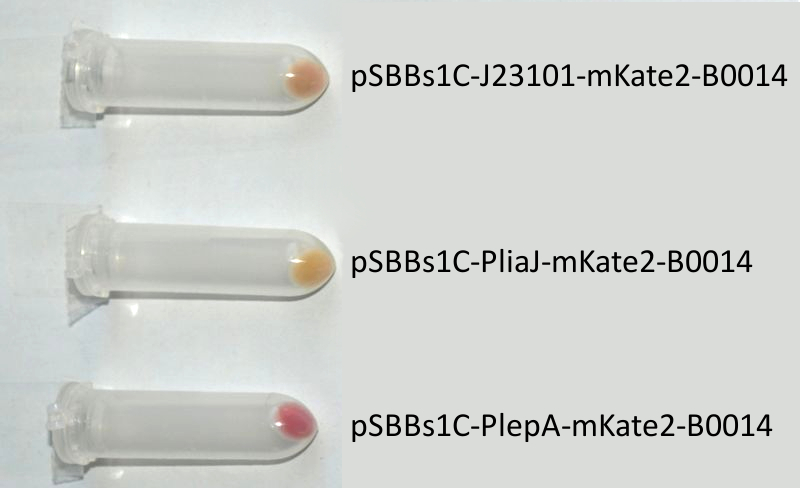
- mKate2 (BioBrick:BBa_K823029)
|
We codon-optimized the sequence of this monomeric far-red fluorescence protein for the use in B. subtilis and synthesized it (by GeneArt) with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation, this reporter was successfully combined with the promoters PliaI, PlepA and the Anderson promoter J23101 in the empty Bacillus vector pSBBs1C from our BacillusBioBrickBox. At the moment we already have the right construct integrated into the chromosome of B. subtilis. Unfortunately, we have not yet measured this reporter.
Reference: mkate2
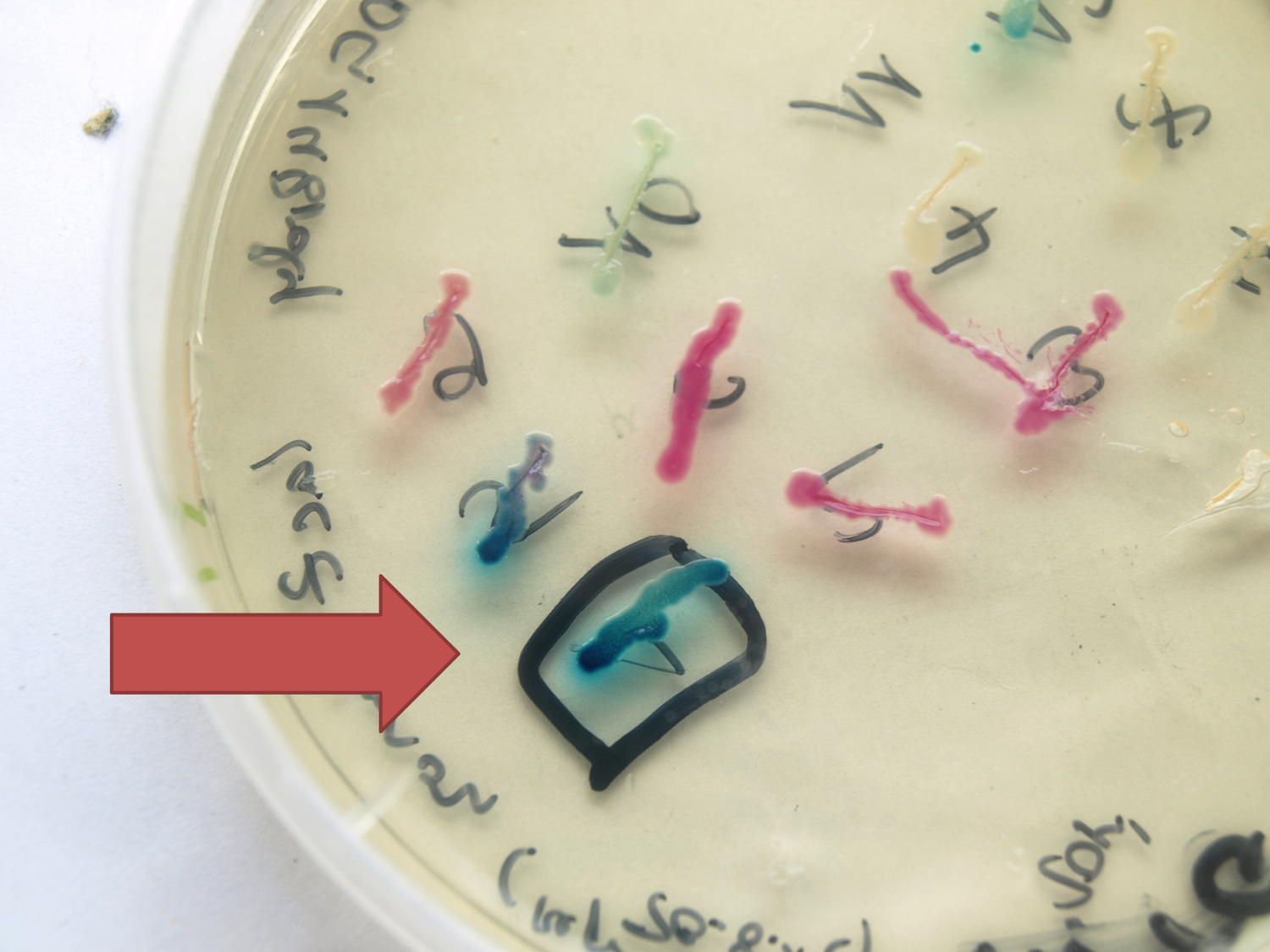
- LacZ (BioBrick:BBa_K823019)
|
This evaluated lacZ gene is derived from the Bacillus reporter vector pAC6. It is constructed in the Freiburg Standard (RFC 25) for in-frame fusion proteins. It also includes a ribosome binding site optimized for Bacillus subtilis translation. This lacZ BioBrick was tested in the expression vector pSBBs0K-Pspac. This construct gave output, so the reporter gene is functional qualitatively. See Data in the vector evaluation section of pSBBs0K-Pspac. A more qualitatively measurement will follow soon.
Reference for the vector: pAC6
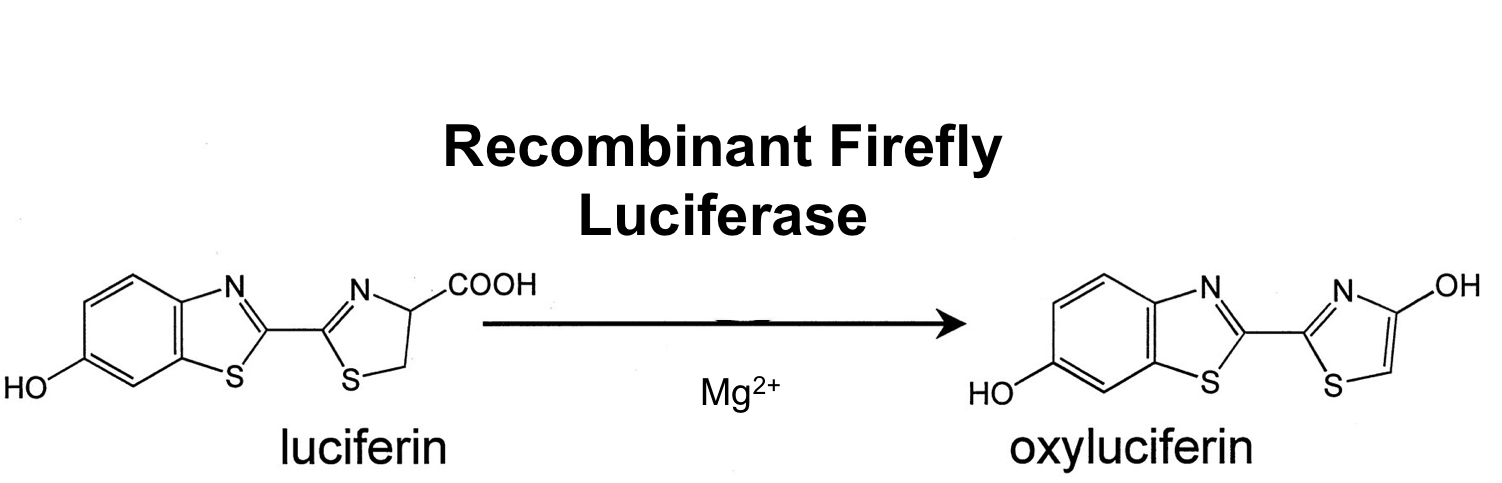
- luc (BioBrick:BBa_K823028)
The luc gene encodes the monomeric firefly luciferase, which requires a special substrate to produce luminescence. We codon-optimized it for Bacillus subtilis, added a ribosome binding site and synthesized by GeneArt.
Firefly luciferase is by far the most commonly used bioluminescent reporter. This monomeric enzyme of 61kDa catalyzes a two-step oxidation reaction to yield light, usually in the green to yellow region, typically 550–570nm . The first step is activation of the luciferyl carboxylate by ATP to yield a reactive mixed anhydride. In the second step, this activated intermediate reacts with oxygen to create a transient dioxetane that breaks down to the oxidized products, oxyluciferin and CO2. Upon mixing with substrates, firefly luciferase produces an initial burst of light that decays over about 15 seconds to a low level of sustained luminescence. This kinetic profile reflects the slow release of the enzymatic product, thus limiting catalytic turnover after the initial reaction.
The popularity of native firefly luciferase as a genetic reporter is due to the sensitivity and convenience of the enzyme assay and tight coupling of protein synthesis with enzyme activity. Firefly luciferase, which is encoded by the luc gene, is a monomer that does not require any post-translational modifications; it is available as a mature enzyme directly upon translation of its mRNA. Catalytic competence is attained immediately after release from the ribosome. Also, luciferase has a very short half-life in cells (approximately 3 hours). Combined, these properties make luciferase an extremely responsive reporter, far more so than other commonly used reporters.
|
It was used in B. subtilis before (Mirouze et al. 2011). The non-optimized version of this reporter gene was already successfully used in B. subtilis. While we have not found the time to evaluate this BioBrick we expect that it will work at least as good.
 "
"