Team:LMU-Munich/Bacillus BioBricks/Promoters
From 2012.igem.org
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
| - | {{:Team:LMU-Munich/Templates/Page Header|File:Team- | + | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_culture_tubes.resized.jpg|3}} |
| + | [[File:Bacillus BioBrick Box banner.resized WORDS.JPG|620px|link=]] | ||
| + | [[Image:BacillusBioBrickBox.png|100px|right|link=]] | ||
| + | ==''Bacillus'' Promoters [[File:LMU PromoterIconBC.png|100px]]== | ||
| + | <p align="justify">To provide a set of promoters of different strength we characterized several promoters in ''Bacillus subtilis''. They can be divided in three different groups: | ||
| + | *the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the [http://partsregistry.org/Main_Page Partsregistry] | ||
| + | *the constitutive promoters P<sub>''liaG''</sub>, P<sub>''veg''</sub> and P<sub>''lepA''</sub> from ''B. subtilis'' | ||
| + | *the inducible promoters P<sub>''liaI''</sub> and ''xylR''-P<sub>''xyl''</sub> from ''B. subtilis''</p> | ||
| + | <p align="justify">For the characterization of the different promoters we used the ''lux'' operon [[File:Lux operon.png|100px]]. Here, promoter activity leads to expression of the luciferase and hence light production, which can be measured as luminescence.</p> | ||
| + | <p align="justify">We also used the reporter gene ''lacZ'' [[File:LacZ.png|50px]]. Here, promoter activation results in expression of a β-galactosidase, whose activity can be measured by breakdown of the chromophoric substrate ONPG (β-galactosidase assays). We used the reporter vector pSB<sub>''Bs''</sub>1C-''lacZ''. See below for an overview and background information of all evaluated promoters and see the [https://2012.igem.org/Team:LMU-Munich/Data Data] page for more details.</p> | ||
| - | |||
| + | ====Overview of all evaluated promoters==== | ||
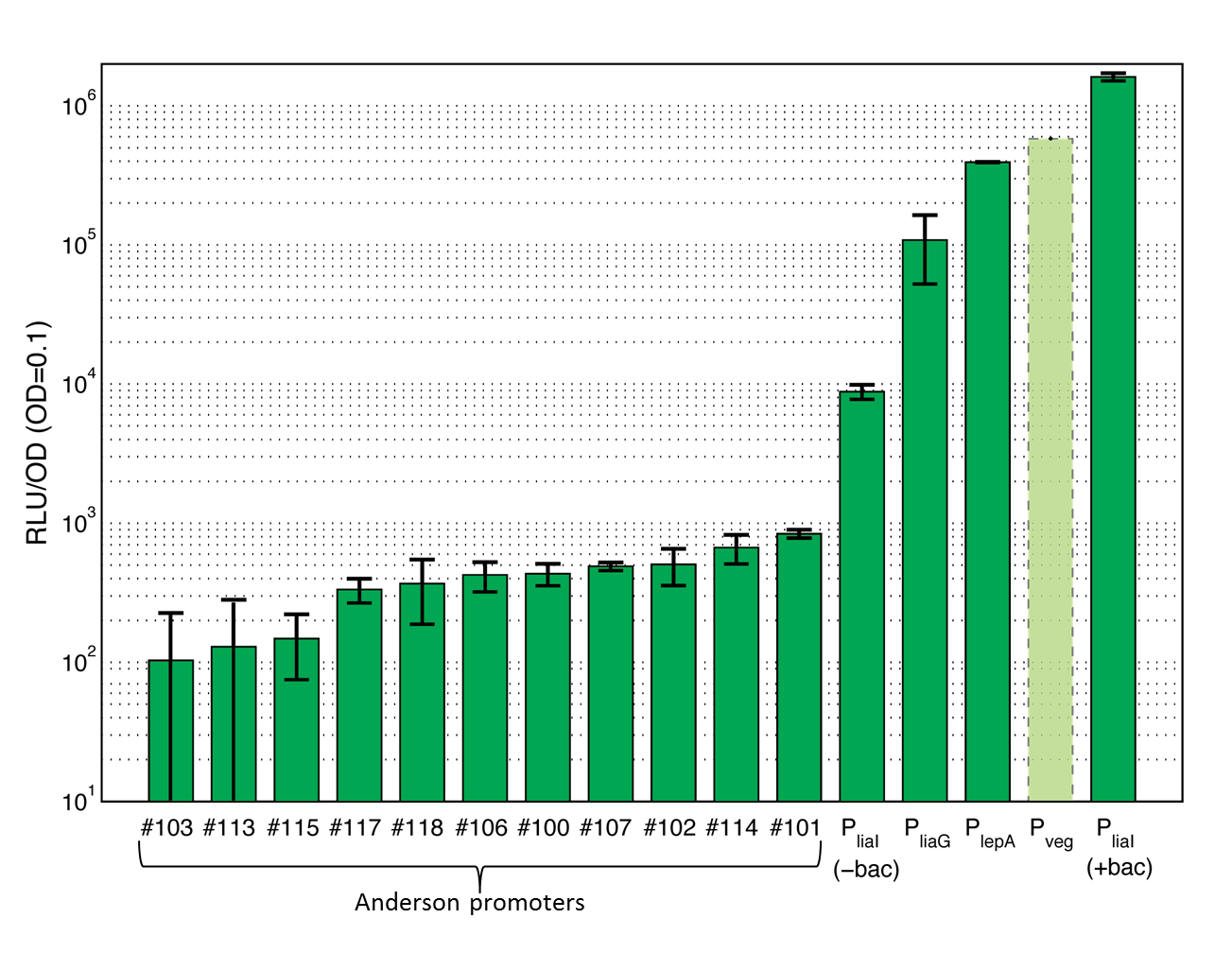
| + | <p align="justify"> This section gives an overview on the strength of all evaluated promoters, which span a large range of activities. For more details and informations of the experiments see the [https://2012.igem.org/Team:LMU-Munich/Data Data] page of the promoters. Note that P<sub>''veg''</sub> was not evaluated with luminescence measurements and this bar is just projected from the results of the β-galactosidase assay.</p> | ||
| + | <br> | ||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:Promoters overview.png|600px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2"><p align="justify"> '''Overview of promoter activity evaluated with luminescence measurements in pSB<sub>''Bs''</sub>3C-''luxABCDE''.''' These values derive from the experiments you can find in our Data section. Lumi per OD<sub>600</sub> are taken at a OD<sub>600</sub> of 0.1. Values are the average and the standard deviation of three different experiments for clone 1. Shown is the activity of the Anderson promoters J23100 (#100), J23101 (#101), J23102 (#102), J23103 (#103), J23106 (#106), J23107 (#107), J23113 (#113), J23114 (#114), J23115 (#115), J23117 (#117), J23118 (#118) as well as the activity of the constitutive promoters P<sub>''liaG''</sub>, and P<sub>''lepA''</sub>. The activity of the inducible promoter P<sub>''liaI''</sub> is shown with (+bac) and without (-bac) induction with bacitracin (10 μg/ml). The promoter activity of P<sub>''veg''</sub> is projected from the results from the β-galactosidase assay and was not measured with luminescence measurements. For the assays shown, an intermediate version of the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' was used, which still contained one forbidden PstI site.</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | ====Evaluation of Anderson promoters in ''B. subtilis''==== | ||
| + | <p align="justify">The first group of promoters evaluated are the promoters of the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] (''"Anderson promoters"''). They have already been extensively measured in ''Escherichia coli'' where they all showed a constitutive behavior with different strength. In this project, eleven Anderson promoters were characterized in ''B. subtilis'' with the ''lux'' operon as a reporter. In ''B. subtilis'' these promoters show quite low activity (see [https://2012.igem.org/Team:LMU-Munich/Data/Anderson#Luminescence_measurements Data Anderson promoters] [[File:Lux operon.png|100px|link=https://2012.igem.org/Team:LMU-Munich/Data/Anderson#Luminescence_measurements]]). | ||
| + | To confirm these results some Anderson promoters were also evaluated with the reporter gene ''lacZ'' by doing β-galactosidase assays (see [https://2012.igem.org/Team:LMU-Munich/Data/Anderson Data Anderson promoters] [[File:LacZ.png|50px|link=https://2012.igem.org/Team:LMU-Munich/Data/Anderson#β-galactosidase_assays]]).</p> | ||
| + | *'''J23100''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823004 BioBrick:BBa_K823004]) | ||
| + | *'''J23101''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823005 BioBrick:BBa_K823005]) | ||
| + | *'''J23102''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823006 BioBrick:BBa_K823006]) | ||
| + | *'''J23103''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823007 BioBrick:BBa_K823007]) | ||
| + | *'''J23106''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823008 BioBrick:BBa_K823008]) | ||
| + | *'''J23107''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823009 BioBrick:BBa_K823009]) | ||
| + | *'''J23113''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823010 BioBrick:BBa_K823010]) | ||
| + | *'''J23114''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823011 BioBrick:BBa_K823011]) | ||
| + | *'''J23115''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823012 BioBrick:BBa_K823012]) | ||
| + | *'''J23117''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823013 BioBrick:BBa_K823013]) | ||
| + | *'''J23118''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823014 BioBrick:BBa_K823014]) | ||
| + | <p align="justify"></p> | ||
| + | ====Constitutive promoters from ''B. subtilis'' as novel BioBricks==== | ||
| + | <p align="justify">The second group of promoters are a set of constitutive promoters from ''B. subtilis'' that we have added to the Registry. We evaluated the promoters [http://partsregistry.org/Part:BBa_K823000 P<sub>''liaG''</sub>], [http://partsregistry.org/Part:BBa_K823003 P<sub>''veg''</sub>] and [http://partsregistry.org/Part:BBa_K823002 P<sub>''lepA''</sub>] using the ''lux'' operon as well as the ''lacZ'' gene as reporters.</p> | ||
| + | |||
| + | *'''P<sub>''liaG''</sub>''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823000 BioBrick:BBa_K823000]) | ||
| + | <p align="justify">P<sub>''liaG''</sub> is a constitutive promoter from ''B. subtilis''. It is responsible for the transcription of the last four genes of the ''liaIHGFSR'' locus and hence for the production of the components of the LiaRS system, which is important for the detection of cell wall antibiotics [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan ''et al.'', 2006)]. P<sub>''liaG''</sub> was evaluated with the ''lux'' operon (see [https://2012.igem.org/Team:LMU-Munich/Data/Constitutive#Luminescence_measurements Data constitutive promoters] [[File:Lux operon.png|100px|link=https://2012.igem.org/Team:LMU-Munich/Data/Constitutive#Luminescence_measurements]]) as well as the ''lacZ'' (see [https://2012.igem.org/Team:LMU-Munich/Data/Constitutive Data constitutive promoters] [[File:LacZ.png|50px|link=https://2012.igem.org/Team:LMU-Munich/Data/Constitutive]]) as reporter. This promoter showed a much higher activity than the Anderson promoters, but was still relatively weak in comparison to other evaluated ''Bacillus'' promoters. </p> | ||
| + | |||
| + | |||
| + | *'''P<sub>''veg''</sub>''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823003 BioBrick:BBa_K823003]) | ||
| + | <p align="justify">P<sub>''veg''</sub> is known to show a strong constitutive activity during the vegetative growth phase and sporulation. This promoter is important for the transcription of the ''veg'' gene, which plays a role during sporulation [http://www.ncbi.nlm.nih.gov/pubmed?term=J.%20Biochem.%2C%20133%20%284%29%3A%20475%E2%80%93483: (Fukushima ''et al.'', 2003)]. P<sub>''veg''</sub> was only measured by using the reporter gene ''lacZ'' (see [https://2012.igem.org/Team:LMU-Munich/Data/Constitutive Data constitutive promoters] [[File:LacZ.png|50px|link=https://2012.igem.org/Team:LMU-Munich/Data/Constitutive]]). This promoter was the strongest of our evaluation. It failed to clone into the ''lux'' reporter vector, presumably because of its strength.</p> | ||
| + | |||
| + | |||
| + | *'''P<sub>''lepA''</sub>''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823002 BioBrick:BBa_K823002]) | ||
| + | <p align="justify"> P<sub>''lepA''</sub> is a constitutive promoter that is important for the transcription of the bicistronic operon. One of the expressed proteins is the protein P<sub>''lepA''</sub> [http://www.ncbi.nlm.nih.gov/pubmed?term=Microbiology%2C%20142%3A%201641%E2%80%931649: (Homuth ''et al.'', 1996)]. P<sub>''lepA''</sub> plays an important role during translation as it can move the mRNA-tRNA complex one step back in the ribosome which is expected to improve the fidelity of translation [http://www.ncbi.nlm.nih.gov/pubmed?term=Cell%2C%20127%20%284%29%3A%20721%E2%80%93733: (Qin ''et al.'', 2006)]. This protein was evaluated with the ''lux'' operon as a reporter (see [https://2012.igem.org/Team:LMU-Munich/Data/Constitutive#Luminescence_measurements Data constitutive promoters] [[File:Lux operon.png|100px|link=https://2012.igem.org/Team:LMU-Munich/Data/Constitutive#Luminescence_measurements]]). The activity of this promoter is between the activity of the strongest ''Bacillus'' promoter P<sub>''veg''</sub> and the weak P<sub>''liaG''</sub>. </p> | ||
| + | |||
| + | |||
| + | ====Inducible promoters from ''B. subtilis'' as a novel BioBrick==== | ||
| + | |||
| + | <p align="justify">The last group of promoters consists of two inducible promoters from ''B. subtilis'', P''<sub>liaI</sub>'' and ''xylR''-P''<sub>xyl</sub>''. They are useful if the strength and timing of gene expression needs to be tightly controlled, because these promoters need an inducer to initiate transcription. P<sub>''liaI''</sub> is evaluated with the reporters ''lux'' and ''lacZ'' and added this novel promoter to the registry.</p> | ||
| + | |||
| + | |||
| + | *'''P<sub>''liaI''</sub>''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823001 BioBrick:BBa_K823001]) | ||
| + | <p align="justify">P''<sub>liaI</sub>'' is an inducible promoter from ''B. subtilis'', which responds to antibiotics that interfere with the integrity and biosynthesis of the cell wall [http://www.ncbi.nlm.nih.gov/pubmed/15273097 (Mascher ''et al.'', 2004)]. In the presence of a stimulus, the two-component system LiaRS is activated. The activated response regulator LiaR binds to the operator of the promoter and induces the transcription of the lia locus. When the promoter is turned on, the two proteins LiaI and LiaH (which play an important role in the stress response of ''B. subtilis'') are expressed. The major strength of this promoter is its very low basal activity in the absence of an inducer and its high dynamic range [http://www.ncbi.nlm.nih.gov/pubmed/15273097 (Mascher ''et al.'', 2004)]. This promoter is evaluated with the reporter ''lux'' (see [https://2012.igem.org/Team:LMU-Munich/Data/Inducible#Luminescence_measurements Data inducible promoters] [[File:Lux operon.png|100px|link=https://2012.igem.org/Team:LMU-Munich/Data/Inducible#Luminescence_measurements]]) as well as ''lacZ'' (see [https://2012.igem.org/Team:LMU-Munich/Data/Inducible Data inducible promoters] [[File:LacZ.png|50px|link=https://2012.igem.org/Team:LMU-Munich/Data/Inducible]]). The induction was measured with different concentrations of bacitracin, demonstrating a concentration-dependent response over a large range of promoter activities.</p> | ||
| + | |||
| + | |||
| + | *'''''xylR''-P<sub>''xyl''</sub>''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K8230015 BioBrick:BBa_K823015]) | ||
| + | <p align="justify">P<sub>''xyl''</sub>-''xylR'' is a xylose-inducible promoter from ''B. subtilis''. XylR is the repressor of P<sub>''xyl''</sub> in the absence of the sugar xylose. In the presence of xylose, XylR dissociates from the operator and P<sub>''xyl''</sub> is active (see e.g. [http://www.ncbi.nlm.nih.gov/pubmed/2544559 Kreuzer et al.]). For this promoter we have not yet suceeded to clone it in a reporter vector to evaluate the activity. So far, there is no data for this promoter.</p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {| width="100%" cellpadding="20" | ||
| + | |[[File:LMU Arrow purple BACK.png|right|80px|link=Team:LMU-Munich/Bacillus_BioBricks]] | ||
| + | |[[File:LMU Arrow purple NEXT.png|left|80px|link=Team:LMU-Munich/Bacillus_BioBricks/Reporters]] | ||
| + | |} | ||
Latest revision as of 12:53, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


Bacillus Promoters 
To provide a set of promoters of different strength we characterized several promoters in Bacillus subtilis. They can be divided in three different groups:
- the constitutive promoters from the Anderson collection from the Partsregistry
- the constitutive promoters PliaG, Pveg and PlepA from B. subtilis
- the inducible promoters PliaI and xylR-Pxyl from B. subtilis
For the characterization of the different promoters we used the lux operon ![]() . Here, promoter activity leads to expression of the luciferase and hence light production, which can be measured as luminescence.
. Here, promoter activity leads to expression of the luciferase and hence light production, which can be measured as luminescence.
We also used the reporter gene lacZ ![]() . Here, promoter activation results in expression of a β-galactosidase, whose activity can be measured by breakdown of the chromophoric substrate ONPG (β-galactosidase assays). We used the reporter vector pSBBs1C-lacZ. See below for an overview and background information of all evaluated promoters and see the Data page for more details.
. Here, promoter activation results in expression of a β-galactosidase, whose activity can be measured by breakdown of the chromophoric substrate ONPG (β-galactosidase assays). We used the reporter vector pSBBs1C-lacZ. See below for an overview and background information of all evaluated promoters and see the Data page for more details.
Overview of all evaluated promoters
This section gives an overview on the strength of all evaluated promoters, which span a large range of activities. For more details and informations of the experiments see the Data page of the promoters. Note that Pveg was not evaluated with luminescence measurements and this bar is just projected from the results of the β-galactosidase assay.
|
Evaluation of Anderson promoters in B. subtilis
The first group of promoters evaluated are the promoters of the Anderson collection ("Anderson promoters"). They have already been extensively measured in Escherichia coli where they all showed a constitutive behavior with different strength. In this project, eleven Anderson promoters were characterized in B. subtilis with the lux operon as a reporter. In B. subtilis these promoters show quite low activity (see Data Anderson promoters ![]() ).
To confirm these results some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters
).
To confirm these results some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters ![]() ).
).
- J23100 (BioBrick:BBa_K823004)
- J23101 (BioBrick:BBa_K823005)
- J23102 (BioBrick:BBa_K823006)
- J23103 (BioBrick:BBa_K823007)
- J23106 (BioBrick:BBa_K823008)
- J23107 (BioBrick:BBa_K823009)
- J23113 (BioBrick:BBa_K823010)
- J23114 (BioBrick:BBa_K823011)
- J23115 (BioBrick:BBa_K823012)
- J23117 (BioBrick:BBa_K823013)
- J23118 (BioBrick:BBa_K823014)
Constitutive promoters from B. subtilis as novel BioBricks
The second group of promoters are a set of constitutive promoters from B. subtilis that we have added to the Registry. We evaluated the promoters PliaG, Pveg and PlepA using the lux operon as well as the lacZ gene as reporters.
- PliaG (BioBrick:BBa_K823000)
PliaG is a constitutive promoter from B. subtilis. It is responsible for the transcription of the last four genes of the liaIHGFSR locus and hence for the production of the components of the LiaRS system, which is important for the detection of cell wall antibiotics (Jordan et al., 2006). PliaG was evaluated with the lux operon (see Data constitutive promoters ![]() ) as well as the lacZ (see Data constitutive promoters
) as well as the lacZ (see Data constitutive promoters ![]() ) as reporter. This promoter showed a much higher activity than the Anderson promoters, but was still relatively weak in comparison to other evaluated Bacillus promoters.
) as reporter. This promoter showed a much higher activity than the Anderson promoters, but was still relatively weak in comparison to other evaluated Bacillus promoters.
- Pveg (BioBrick:BBa_K823003)
Pveg is known to show a strong constitutive activity during the vegetative growth phase and sporulation. This promoter is important for the transcription of the veg gene, which plays a role during sporulation (Fukushima et al., 2003). Pveg was only measured by using the reporter gene lacZ (see Data constitutive promoters ![]() ). This promoter was the strongest of our evaluation. It failed to clone into the lux reporter vector, presumably because of its strength.
). This promoter was the strongest of our evaluation. It failed to clone into the lux reporter vector, presumably because of its strength.
- PlepA (BioBrick:BBa_K823002)
PlepA is a constitutive promoter that is important for the transcription of the bicistronic operon. One of the expressed proteins is the protein PlepA (Homuth et al., 1996). PlepA plays an important role during translation as it can move the mRNA-tRNA complex one step back in the ribosome which is expected to improve the fidelity of translation (Qin et al., 2006). This protein was evaluated with the lux operon as a reporter (see Data constitutive promoters ![]() ). The activity of this promoter is between the activity of the strongest Bacillus promoter Pveg and the weak PliaG.
). The activity of this promoter is between the activity of the strongest Bacillus promoter Pveg and the weak PliaG.
Inducible promoters from B. subtilis as a novel BioBrick
The last group of promoters consists of two inducible promoters from B. subtilis, PliaI and xylR-Pxyl. They are useful if the strength and timing of gene expression needs to be tightly controlled, because these promoters need an inducer to initiate transcription. PliaI is evaluated with the reporters lux and lacZ and added this novel promoter to the registry.
- PliaI (BioBrick:BBa_K823001)
PliaI is an inducible promoter from B. subtilis, which responds to antibiotics that interfere with the integrity and biosynthesis of the cell wall (Mascher et al., 2004). In the presence of a stimulus, the two-component system LiaRS is activated. The activated response regulator LiaR binds to the operator of the promoter and induces the transcription of the lia locus. When the promoter is turned on, the two proteins LiaI and LiaH (which play an important role in the stress response of B. subtilis) are expressed. The major strength of this promoter is its very low basal activity in the absence of an inducer and its high dynamic range (Mascher et al., 2004). This promoter is evaluated with the reporter lux (see Data inducible promoters ![]() ) as well as lacZ (see Data inducible promoters
) as well as lacZ (see Data inducible promoters ![]() ). The induction was measured with different concentrations of bacitracin, demonstrating a concentration-dependent response over a large range of promoter activities.
). The induction was measured with different concentrations of bacitracin, demonstrating a concentration-dependent response over a large range of promoter activities.
- xylR-Pxyl (BioBrick:BBa_K823015)
Pxyl-xylR is a xylose-inducible promoter from B. subtilis. XylR is the repressor of Pxyl in the absence of the sugar xylose. In the presence of xylose, XylR dissociates from the operator and Pxyl is active (see e.g. Kreuzer et al.). For this promoter we have not yet suceeded to clone it in a reporter vector to evaluate the activity. So far, there is no data for this promoter.
 "
"