Team:LMU-Munich/Bacillus BioBricks
From 2012.igem.org
| Line 192: | Line 192: | ||
==''Bacillus'' Promoters [[File:LMU PromoterIconBC.png|100px]]== | ==''Bacillus'' Promoters [[File:LMU PromoterIconBC.png|100px]]== | ||
| - | <p align="justify">To get a set of promoters with different strength we characterized several promoters in ''Bacillus subtilis''. They can be divided in three different groups: the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Partsregistry, the constitutive promoters P<sub>''liaG''</sub>, P<sub>''veg''</sub> and P<sub>''lepA''</sub> from ''B. subtilis'', and the inducible promoters P<sub>''liaI''</sub> and P<sub>''xyl''</sub>-''XylR'' from ''B. subtilis''. For the characterization of the different promoters we used the ''lux'' operon where gene expression leads to the production of luminescence | + | <p align="justify">To get a set of promoters with different strength we characterized several promoters in ''Bacillus subtilis''. They can be divided in three different groups: the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Partsregistry, the constitutive promoters P<sub>''liaG''</sub>, P<sub>''veg''</sub> and P<sub>''lepA''</sub> from ''B. subtilis'', and the inducible promoters P<sub>''liaI''</sub> and P<sub>''xyl''</sub>-''XylR'' from ''B. subtilis''. For the characterization of the different promoters we used the ''lux'' operon where gene expression leads to the production of luminescence. For this promoter evaluation the reporter vector pSB<sub>''Bs''</sub>3C-''luxABCDE'' was used which was not fully in BioBrickStandard in this time because of one last forbidden restriction site. We also used the reporter gene ''lacZ'' for the promoter evaluation where the promoter activity can be measured by doing β-galactosidase assays. Therefore we used the reporter vector pSB<sub>''Bs''</sub>1C-''lacZ''.</p> |
| Line 259: | Line 259: | ||
*'''mKate''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029]) | *'''mKate''' ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029]) | ||
| - | <p align="justify">We synthesized this ''rfp'' derivate with a codon-optimized version for the use in ''B. subtilis'' with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation this reporter was successfully combined with the promoters PlepA, PliaI and J23101 in the empty Bacillus vector pSB''Bs''1C from our '''''Bacillus''B'''io'''B'''rick'''B'''ox. </p> | + | <p align="justify">We synthesized this ''rfp'' derivate with a codon-optimized version for the use in ''B. subtilis'' with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation this reporter was successfully combined with the promoters PlepA, PliaI and J23101 in the empty ''Bacillus'' vector pSB<sub>''Bs''</sub>1C from our '''''Bacillus''B'''io'''B'''rick'''B'''ox. </p> |
| Line 266: | Line 266: | ||
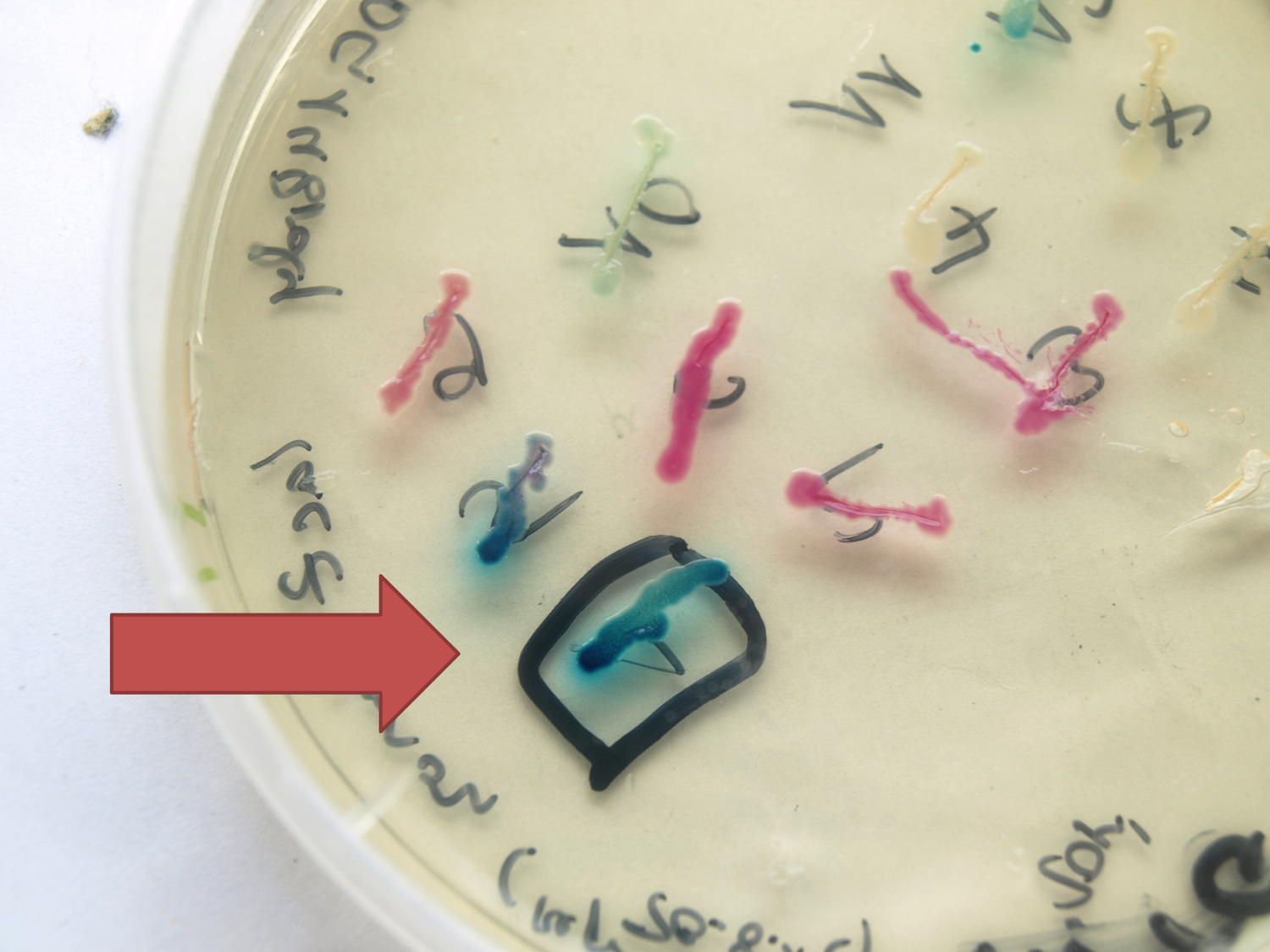
[[File:LacZ plate.png|''lacZ'' under the control of P<sub>''spac''</sub> in pSB<sub>Bs</sub>0K-P<sub>''spac''</sub>. Plate induced with IPTG|thumb|300px|left]] | [[File:LacZ plate.png|''lacZ'' under the control of P<sub>''spac''</sub> in pSB<sub>Bs</sub>0K-P<sub>''spac''</sub>. Plate induced with IPTG|thumb|300px|left]] | ||
| - | <p align="justify">This lacZ gene is derived from the ''Bacillus'' reporter vector pAC6. It is constructed in the Freiburg Standard (Assembly 25) for in-frame fusion proteins. It also includes a Shine-Dalgarno Sequence optimized for ''Bacillus subtilis'' translation.</p> | + | <p align="justify">This ''lacZ'' gene is derived from the ''Bacillus'' reporter vector pAC6. It is constructed in the Freiburg Standard (Assembly 25) for in-frame fusion proteins. It also includes a Shine-Dalgarno Sequence optimized for ''Bacillus subtilis'' translation. This ''lacZ'' BioBrick was tested with the promoters P<sub>''liaG''</sub> and P<sub>''veg''</sub> and the results were directly compared to the ''lacZ'' from our reporter vector pSB<sub>''Bs''</sub>1C-''lacZ''. </p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 17:23, 23 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


Achtung: In Bearbeitung
BacillusBioBrickBox
We will create a toolbox of Bacillus BioBricks to contribute to the registry.
This BacillusBioBrickBox contains Bacillus specific:
| Vectors | Promoters | Reporters | Affinity tags |

| 
| 
| |
|
pSBBs1C |
Pveg |
mkate2 |
Flag |
Bacillus Vectors 
Here is a list of all the vectors we cloned and used. Please note that pMAD is not in a BioBrick standard, but it is a useful tool to knock out genes in Bacillus subtilis.
For the use of our vectors, please see our Protocols page. A general introduction to Bacillus subtilis and its integrative vectors can be found here. All vectors have ampicillin as E. coli resistance and RFP in the multiple cloning site as selection marker.
| Name BioBrick | E. coli res. | B. subt. res. | Insertion locus | Description | Derived from | Reference |
|---|---|---|---|---|---|---|
| pSBBs1C [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823023 (BBa_K823023)] | Amp | Cm | amyE | empty | pDG1662 | [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury et al.] |
| pSBBs4S [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823022 (BBa_K823022)] | Amp | Spec | thrC | empty | pDG1731 | [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury et al.] |
| pSBBs2E [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823027 (BBa_K823027)] | Amp | MLS | lacA | empty | pAX01 | [http://www.ncbi.nlm.nih.gov/pubmed/11274134 Härtl et al.] |
| pSBBs1C-lacZ [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823021 (BBa_K823021) ] | Amp | Cm | amyE | with lacZ reporter gene | pAC6 | [http://www.ncbi.nlm.nih.gov/pubmed/11902727 Stülke et al.] |
| pSBBs3C-luxABCDE [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 (BBa_K823025)] | Amp | Cm | sacA | with luxABCDE reporter cassette | pAH328 | [http://www.ncbi.nlm.nih.gov/pubmed/20709900 Schmalisch et al.] |
| pSBBs4S-PXyl [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823024 (BBa_K823024)] | Amp | Spec | thrC | with Xylose-inducible promoter | pXT | [http://www.ncbi.nlm.nih.gov/pubmed/11069659 Derré et al.] |
| pSBBs0K-Pspac [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 (BBa_K823026)] | Amp | Kan | replicative | with IPTG inducible promoter | pDG148 | [http://www.ncbi.nlm.nih.gov/pubmed/11728721 Joseph et al.] |
| Sporovector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823054 (BBa_K823054)] | Amp | Spec | thrC | to create Sporobeads | pSBBs4S |
Here you can find the respective vector maps:

| 
|

| 
|

| 
|

| 
|
The number in the vector's name codes for the insertion locus and the following letter for the Bacillus subtilis resistance gene according to the following table:
| Number | Insertion locus | Letter | Resistance |
|---|---|---|---|
| 0 | replicative | C | Chloramphenicol |
| 1 | amyE (amylase) | E | MLS (Erythromycin + Lincomycin) |
| 2 | lacA (β-galactosidase) | K | Kanamycin |
| 3 | sacA (sucrase) | S | Spectinomycin |
| 4 | thrC (threonine C) |
The concentrations of the antibiotics and the insertion tests can be found in our Protocol section.
Bacillus Promoters 
To get a set of promoters with different strength we characterized several promoters in Bacillus subtilis. They can be divided in three different groups: the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Partsregistry, the constitutive promoters PliaG, Pveg and PlepA from B. subtilis, and the inducible promoters PliaI and Pxyl-XylR from B. subtilis. For the characterization of the different promoters we used the lux operon where gene expression leads to the production of luminescence. For this promoter evaluation the reporter vector pSBBs3C-luxABCDE was used which was not fully in BioBrickStandard in this time because of one last forbidden restriction site. We also used the reporter gene lacZ for the promoter evaluation where the promoter activity can be measured by doing β-galactosidase assays. Therefore we used the reporter vector pSBBs1C-lacZ.
Anderson promoters
The first group of promoters evaluated are the promoters of the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] which we call Anderson promoters. They have already been measured in Escherichia coli where they all showed a constitutive behavior with a different strength. In this project, eleven Anderson promoters were characterized in B. subtilis with the lux operon as a reporter. In this evaluation in B. subtilis they showed quiet a low activity (see Data Anderson promoters ![]() ). To confirm this result some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters
). To confirm this result some Anderson promoters were also evaluated with the reporter gene lacZ by doing β-galactosidase assays (see Data Anderson promoters ![]() ).
).
- J23100 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823004 BioBrick:BBa_K823004])
- J23101 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823005 BioBrick:BBa_K823005])
- J23102 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823006 BioBrick:BBa_K823006])
- J23103 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823007 BioBrick:BBa_K823007])
- J23106 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823008 BioBrick:BBa_K823008])
- J23107 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823009 BioBrick:BBa_K823009])
- J23113 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823010 BioBrick:BBa_K823010])
- J23114 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823011 BioBrick:BBa_K823011])
- J23115 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823012 BioBrick:BBa_K823012])
- J23117 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823013 BioBrick:BBa_K823013])
- J23118 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823014 BioBrick:BBa_K823014])
Constitutive promoters from B. subtilis
The second group of promoters charaterized are the constitutive promoters from B. subtilis. We evaluated the promoters PliaG, Pveg and PlepA. Therefore we used the lux operon as reporter as well as the lacZ gene.
- PliaG ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823000 BioBrick:BBa_K823000])
PliaG is a weak, constitutive promoter from B. subtilis. It is responsible for the transcription of the last four genes of the liaIHGFSR locus and therefore for the production of the components of the LiaRS system, which is important for the detection of cell wall antibiotics [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan et al., 2006)]. PliaG was evaluated with the lux operon as reporter (see Data constitutive promoters ![]() ) and the reporter lacZ (see Data constitutive promoters
) and the reporter lacZ (see Data constitutive promoters ![]() ). This promoter showed a much higher activity than the Anderson promoters which was still weak in comparison to the other evaluated Bacillus promoters.
). This promoter showed a much higher activity than the Anderson promoters which was still weak in comparison to the other evaluated Bacillus promoters.
- Pveg ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823003 BioBrick:BBa_K823003])
Pveg is known to show a strong constitutive activity during the vegetative growth phase and sporulation phase. This promoter is important for the transcription of the veg gene, which plays an important role during sporulation [http://www.ncbi.nlm.nih.gov/pubmed?term=J.%20Biochem.%2C%20133%20%284%29%3A%20475%E2%80%93483: (Fukushima et al., 2003)]. Pveg was measured by using reporter gene lacZ (see Data constitutive promoters ![]() ). This promoter was the strongest of our evaluated promoters.
). This promoter was the strongest of our evaluated promoters.
- PlepA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823002 BioBrick:BBa_K823002])
PlepA is constitutive promoter which is important for the transcription of a bicistronic operon. One of the expressed proteins is the protein PlepA [http://www.ncbi.nlm.nih.gov/pubmed?term=Microbiology%2C%20142%3A%201641%E2%80%931649: (Homuth et al., 1996)]. This protein plays an important role during the translation as it can move the mRNA-tRNA complex one step back in the ribosome which is expected to improve the fidelity of translation [http://www.ncbi.nlm.nih.gov/pubmed?term=Cell%2C%20127%20%284%29%3A%20721%E2%80%93733: (Qin et al., 2006)]. This promoter was evaluated with the lux operon as a reporter (see Data constitutive promoters ![]() ). The activity of this promoter is between the activity of the strongest promoter Pveg and the weak Bacillus promoter PliaG.
). The activity of this promoter is between the activity of the strongest promoter Pveg and the weak Bacillus promoter PliaG.
Inducible promoters from B. subtilis
The last group of promoters consists of two inducible promoters of B. subtilis , PliaI and Pxyl-XylR. They are useful to decide when to turn on gene expression because these promoters need an inducer to start transcription. They are evaluated with the reporters lux and lacZ.
- PliaI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823001 BioBrick:BBa_K823001])
PliaI is an inducible promoter from B. subtilis which is induced by antibiotics that interact with the lipidII cycle, e.g. bacitracin. If the cell senses stress there is a phosphorylation of the two proteins LiaS and LiaR. LiaR binds to the operator of the promoter and induces the transcription. When the promoter is turned on the two proteins LiaI and LiaH are expressed which play an important role in the stress response [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan et al., 2006)]. This promoter is evaluated with the reporters lux (see Data inducible promoters ![]() ) as well as lacZ (see Data inducible promoters
) as well as lacZ (see Data inducible promoters ![]() ). The induction was measured with different concentrations of bacitracin. With induction with different concentrations of bacitracin this promoter covers a large range of activity.
). The induction was measured with different concentrations of bacitracin. With induction with different concentrations of bacitracin this promoter covers a large range of activity.
- PXyl-XylR ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K8230015 BioBrick:BBa_K823015])
PXyl-XylR is a inducible promoter from B. subtilis which is induced by xylose. XylR is constitutively expressed, and is the repressor of PXyl in absence of the sugar Xylose. In presence of Xylose, XylR leaves the operator and the PXyl is active [see e.g. [http://www.ncbi.nlm.nih.gov/pubmed/2544559 Kreuzer et al.]. For this promoter we have not yet suceeded cloning it in the reporter vector to evaluate the activity. So there is not any data for this promoter yet.
Bacillus Reporters 
We designed some reporters that are commonly used in B. subtilis or are codon optimized versions of popular reporter genes. All reporters have a modified iGEM Freiburg standard ([http://partsregistry.org/Help:Assembly_standard_25 RCF 25]) pre- and suffix for assembly of in-frame fusion proteins. Our prefix also includes the B. subitlis optimized RBS.
prefix: GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix: ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
- GFP ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823039 BioBrick:BBa_K823039])
We took a gfp derivate of the Bacillus subtilis plasmids pGFPamy and added the BioBrick compatiple pre- and suffix of the Freiburg standard (Assembly 25).
- mKate ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823029 BioBrick:BBa_K823029])
We synthesized this rfp derivate with a codon-optimized version for the use in B. subtilis with pre- and suffix of the Freiburg standard. We cloned this reporter in front of the terminator B0014. For the evaluation this reporter was successfully combined with the promoters PlepA, PliaI and J23101 in the empty Bacillus vector pSBBs1C from our BacillusBioBrickBox.
- LacZ ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823019 BioBrick:BBa_K823019])
This lacZ gene is derived from the Bacillus reporter vector pAC6. It is constructed in the Freiburg Standard (Assembly 25) for in-frame fusion proteins. It also includes a Shine-Dalgarno Sequence optimized for Bacillus subtilis translation. This lacZ BioBrick was tested with the promoters PliaG and Pveg and the results were directly compared to the lacZ from our reporter vector pSBBs1C-lacZ.
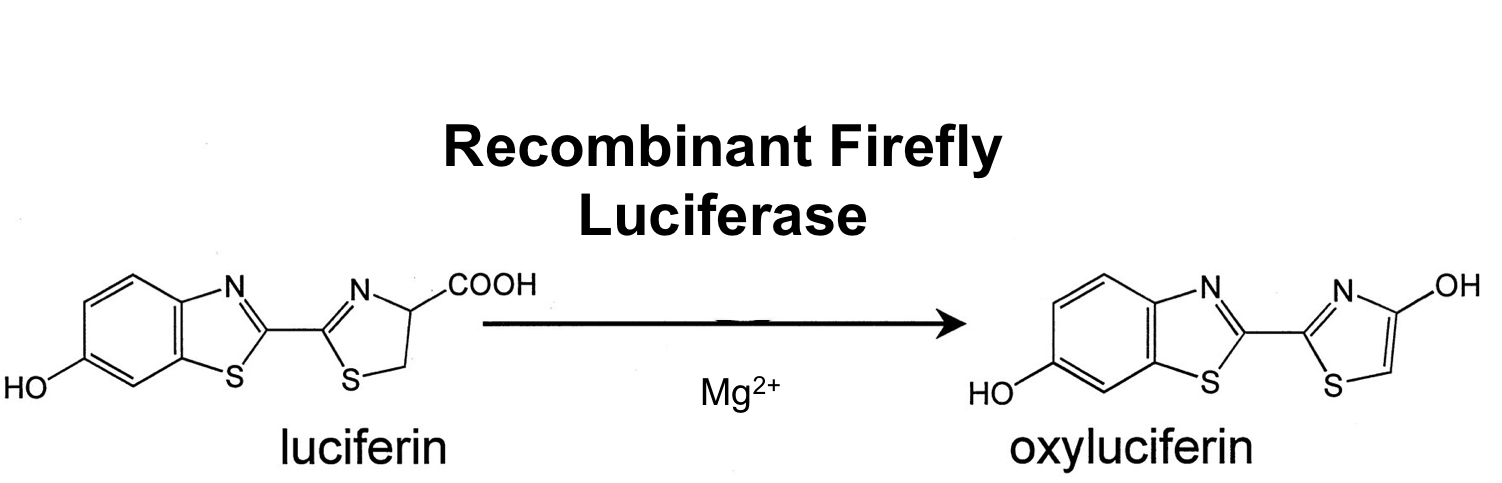
- luc ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823028 BioBrick:BBa_K823028])
We synthesized this luciferase for luminescence assays with luciferin as substrate.
Affinity Tags 
- 3x Flag - tag [http://partsregistry.org/Part:BBa_K823034 (BioBrick:BBa_K823034)]
- HA - tag [http://partsregistry.org/Part:BBa_K823035 (BioBrick:BBa_K823035)]
- cMyc - tag [http://partsregistry.org/Part:BBa_K823036 (BioBrick:BBa_K823036)]
- His - tag [http://partsregistry.org/Part:BBa_K823037 (BioBrick:BBa_K823037)]
- Strep - tag [http://partsregistry.org/Part:BBa_K823038 (BioBrick:BBa_K823038)]

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"