Team:LMU-Munich/Application/Protein Screening tour
From 2012.igem.org
| (One intermediate revision not shown) | |||
| Line 8: | Line 8: | ||
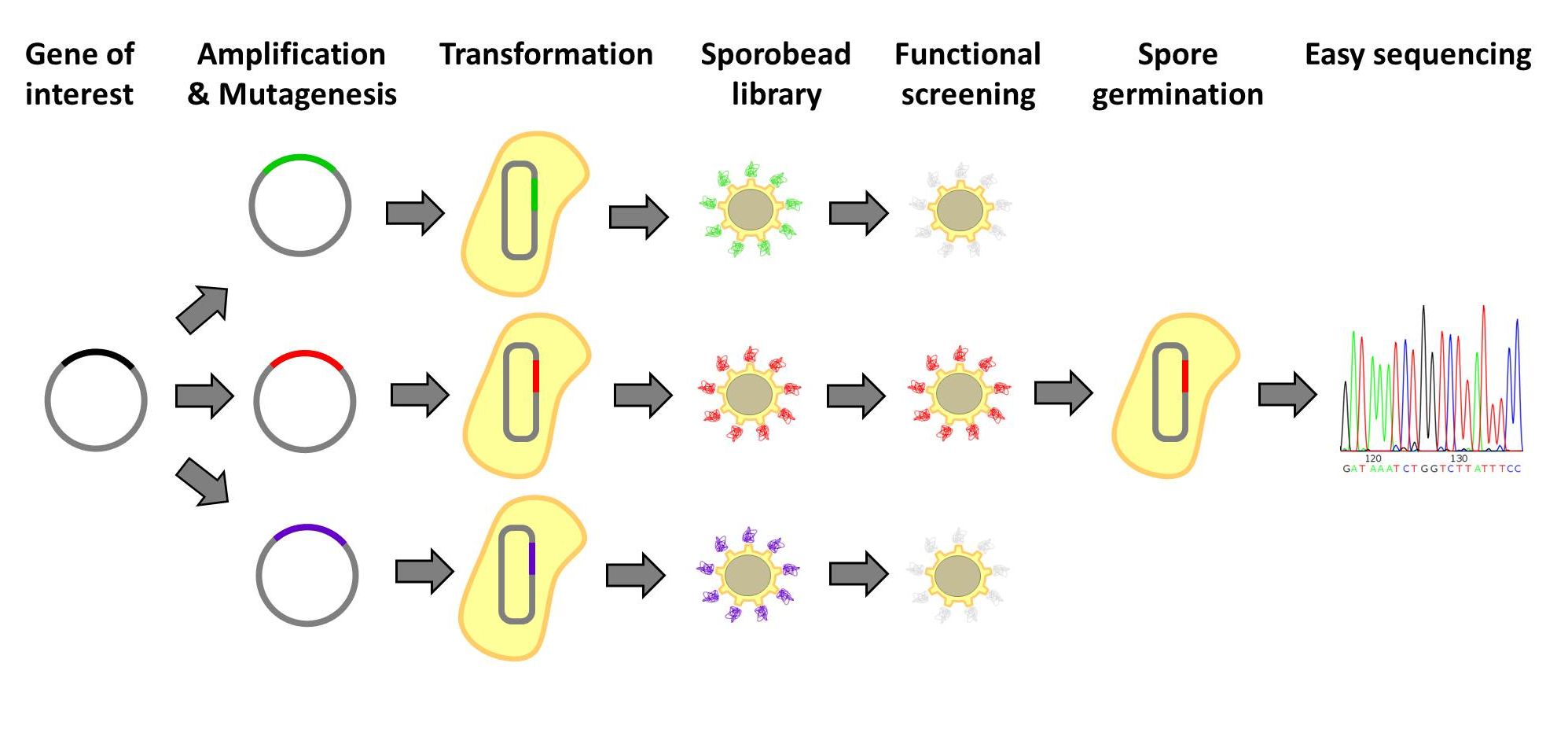
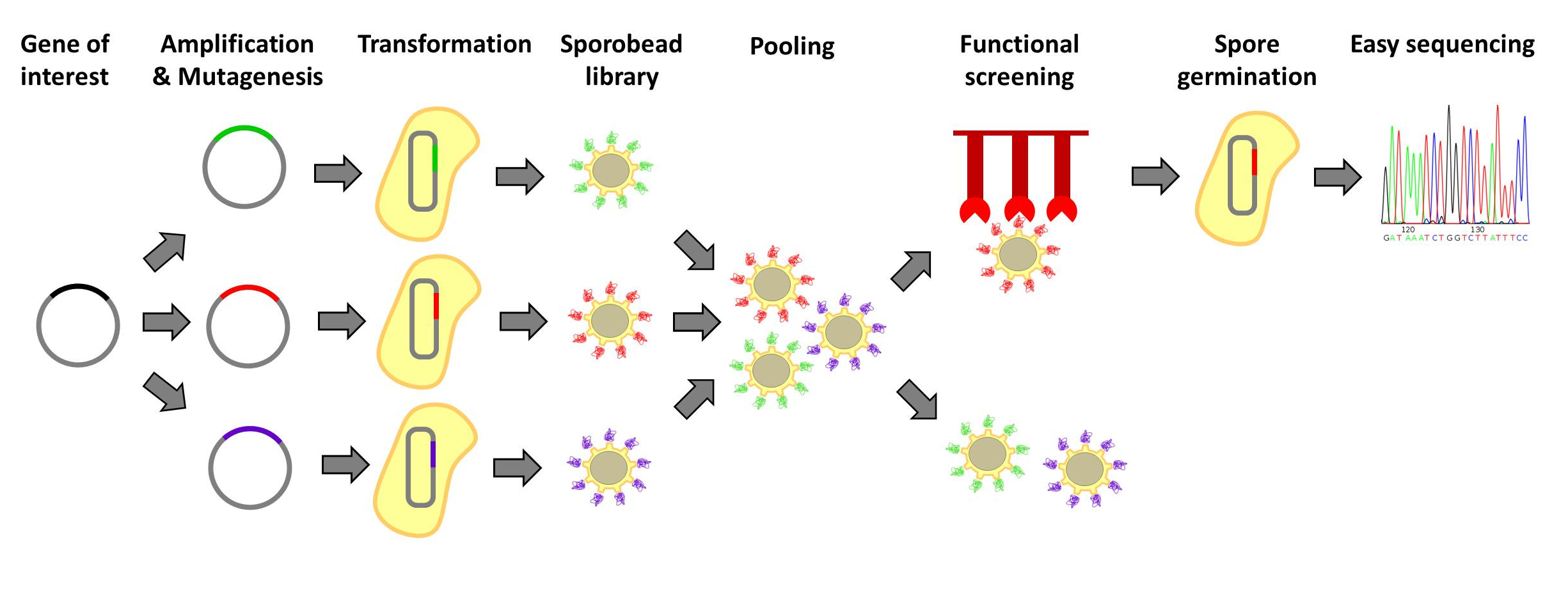
| - | <p align="justify">[http://www.ncbi.nlm.nih.gov/ | + | <p align="justify">[http://www.ncbi.nlm.nih.gov/pubmed/20038141 Designer proteins], which enhance binding capacities or improve enzymatic activities, have a large potential in both research and biotechnological processes. They are usually created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in functional screenings. Despite the success of the method, it is labor intensive and limited in throughput. First protein extracts need to be prepared. Second, after successful screens individual mutated proteins must be tracked back to the original strains. Our '''Sporo'''beads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. Figures 1 and 2 offer schematics of the process of using '''Sporo'''beads for protein screening for either enzymatic or binding affinities.</p> |
| Line 14: | Line 14: | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
{|align:center | {|align:center | ||
| - | |[[File:protein_libraries.jpg|620px|center]] | + | |[[File:protein_libraries.jpg|620px|center|link=]] |
|- | |- | ||
| style="width: 80%;background-color: #EBFCE4;" | | | style="width: 80%;background-color: #EBFCE4;" | | ||
| Line 27: | Line 27: | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
{|align:center | {|align:center | ||
| - | |[[File:protein_libraries_affinities.jpg|620px|center]] | + | |[[File:protein_libraries_affinities.jpg|620px|center|link=]] |
|- | |- | ||
| style="width: 80%;background-color: #EBFCE4;" | | | style="width: 80%;background-color: #EBFCE4;" | | ||
Latest revision as of 17:12, 26 October 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


Protein Screening
Designer proteins, which enhance binding capacities or improve enzymatic activities, have a large potential in both research and biotechnological processes. They are usually created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in functional screenings. Despite the success of the method, it is labor intensive and limited in throughput. First protein extracts need to be prepared. Second, after successful screens individual mutated proteins must be tracked back to the original strains. Our Sporobeads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. Figures 1 and 2 offer schematics of the process of using Sporobeads for protein screening for either enzymatic or binding affinities.
|
|
 "
"