|
|
| (5 intermediate revisions not shown) |
| Line 34: |
Line 34: |
| | {| width="100%" style="text-align:center;"| | | {| width="100%" style="text-align:center;"| |
| | |<p align="justify">Further information for the first possible application for our '''Sporo'''beads.</p> | | |<p align="justify">Further information for the first possible application for our '''Sporo'''beads.</p> |
| - | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Application/Filtration]] | + | |[[File:LMU Sporofilter.png|right|150px|link=Team:LMU-Munich/Application/Filtration]] |
| | |- | | |- |
| | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Filtration]] | | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Filtration]] |
| | |} | | |} |
| | </div> | | </div> |
| - |
| |
| - | ==Filtration==
| |
| - |
| |
| - | <p align="justify">Our '''Sporo'''beads can express proteins on their outer coats to bind specific molecular targets. Such targets include heavy metals, toxins, and plastic.</p>
| |
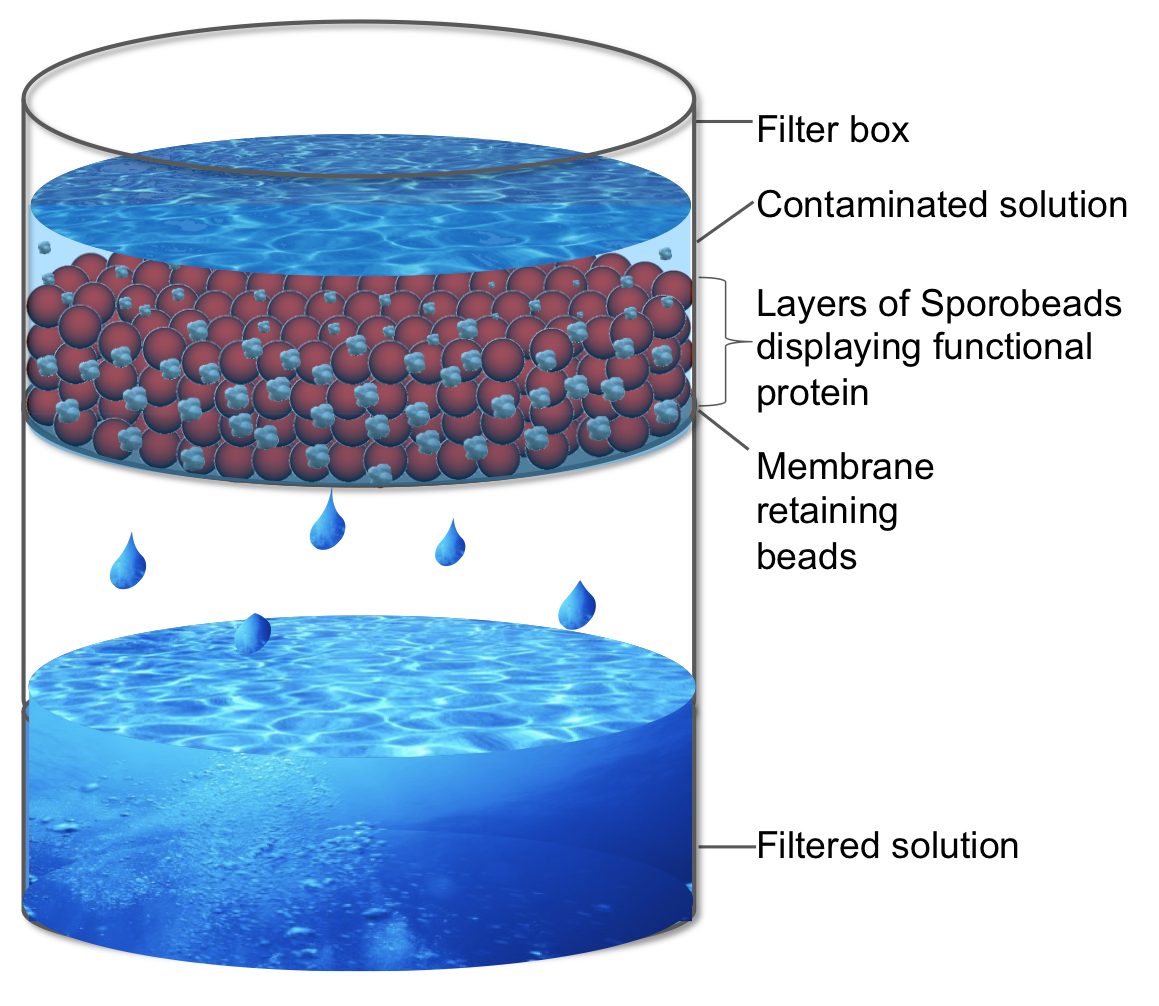
| - | <p align="justify">One example of such a filtering protein could be a CPX-'''Sporo'''bead. [http://partsregistry.org/wiki/index.php/Part:BBa_I728500 CPX] is a peptide developed by the [https://2007.igem.org/MIT 2007 MIT iGEM team], which is extremely hydrophilic, and thus capable of binding microparticles of polystyrene from water. The excessive use of disposable plastic and the lack of universal recycling programs has led to the [http://www.ncbi.nlm.nih.gov/pubmed/22610295 pollution of the world's oceans]. In the ocean, large pieces of plastic litter are ground by sea currents and degraded by UV radiation into microscopic pieces, so called "plastic plankton," which is consumed by fish, filter feeders, and other marine organisms. Such plastic uptake can lead to poisoning, sterility and death.</p>
| |
| - | <p align="justify">CPX-'''Sporo'''beads in huge filter boxes could be put into place to mechanically filter microscopic plastic particles out of the water. Such specific filtration would be superior to blanket filtration systems, which also remove living phytoplankton important to ocean ecosystems. To prevent the beads from being released into the sea and to ensure the plastic be removed from the water, the '''Sporo'''beads could be attached to membranes in the filter boxes. Then the '''Sporo'''beads would need to not only display CPX but also a membrane binding protein on their surface.</p>
| |
| | | | |
| | <div class="box"> | | <div class="box"> |
| Line 50: |
Line 44: |
| | {| width="100%" style="text-align:center;"| | | {| width="100%" style="text-align:center;"| |
| | |<p align="justify">Further information for the possible application of protein screening.</p> | | |<p align="justify">Further information for the possible application of protein screening.</p> |
| - | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Application/Protein Screening]] | + | |[[File:protein_libraries.jpg|right|150px|link=Team:LMU-Munich/Application/Protein Screening]] |
| | |- | | |- |
| | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Protein Screening]] | | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Protein Screening]] |
| | |} | | |} |
| | </div> | | </div> |
| - |
| |
| - | ==Protein Screening==
| |
| - |
| |
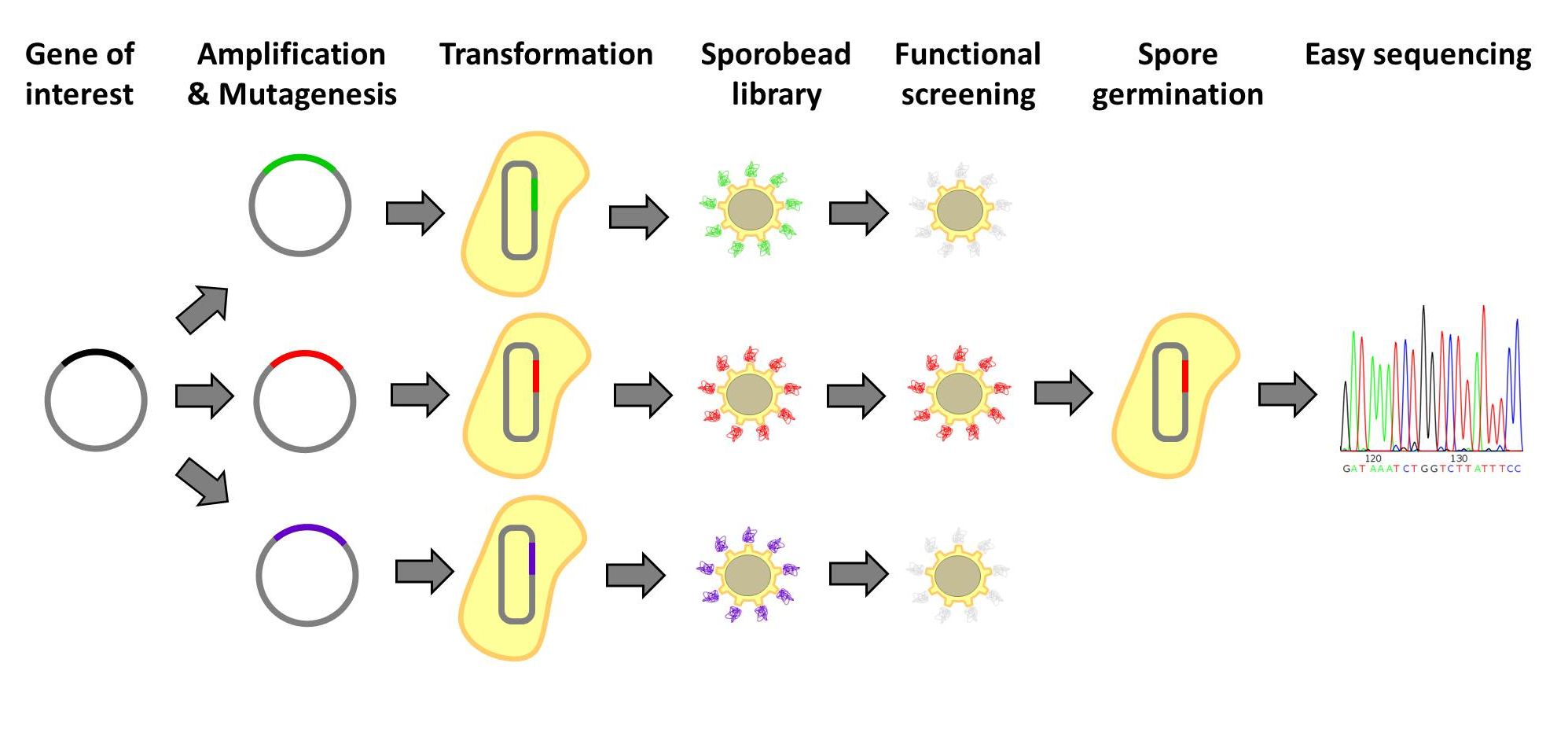
| - | <p align="justify">Designer protein molecules, which bind their desired targets specifically, have numerous uses. Designer proteins are frequently created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in this method. Despite the success of the method, it is labor intensive and limited in throughput. Individual mutated proteins must be tracked throughout the entire screening process. Our '''Sporo'''beads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. Figures 1 and 2 offer schematics of the process of using '''Sporo'''beads for protein screening for tests and affinities.</p>
| |
| - |
| |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {|align:center
| |
| - | |[[File:protein_libraries.jpg|620px|center]]
| |
| - | |-
| |
| - | | style="width: 80%;background-color: #EBFCE4;" |
| |
| - | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;"
| |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| |
| - | <font color="#000000"; size="2">Fig. 1: '''The simple protein-screening process in our Sporobeads'''. </font>
| |
| - | |}
| |
| - | |}
| |
| - | |}
| |
| - |
| |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {|align:center
| |
| - | |[[File:protein_libraries_affinities.jpg|620px|center]]
| |
| - | |-
| |
| - | | style="width: 80%;background-color: #EBFCE4;" |
| |
| - | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;"
| |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| |
| - | <font color="#000000"; size="2">Fig. 2: '''The simple protein-screening process in our Sporobeads''' using affinity binding as a requisite. </font>
| |
| - | |}
| |
| - | |}
| |
| - | |}
| |
| - | <br>
| |
| - |
| |
| | | | |
| | <div class="box"> | | <div class="box"> |
| | ====Further Applications==== | | ====Further Applications==== |
| | {| width="100%" style="text-align:center;"| | | {| width="100%" style="text-align:center;"| |
| - | |<p align="justify">Further information for the possible application of protein screening.</p> | + | |<p align="justify">More information for further possible application with our diverse '''Sporo'''beads.</p> |
| - | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Application/Further Applications]] | + | |[[File:LMU Sporo diversity.png|right|150px|link=Team:LMU-Munich/Application/Further Applications]] |
| | |- | | |- |
| | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Further Applications]] | | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Application/Further Applications]] |
| | |} | | |} |
| | </div> | | </div> |
| - | | + | <br> |
| - | ==Further Applications==
| + | <br> |
| - | | + | <br> |
| - | <p align="justify">Ultimately, the ability to express virtually any protein of interest shows great potential for laboratory work. Our '''Sporo'''beads could additionally express TAL effectors, for the binding of sequence-specific DNA stretches. This could allow simple GMO detection of food crops. Besides simply expressing proteins capable of binding elements of interest, our '''Sporo'''beads could express proteins which have enzymatic activity. The 2011 University of Washington iGEM team developed an enzyme, Kumamolisin, which cleaves peptides. The specific substrate of this enzyme is the specific amino acid sequence which causes reactions in individuals with Celiac disease. Such a cleaving enzyme could be used to eliminate irritants from gluten-containing food products.</p> | + | <br> |
| - | | + | |
| - | <p align="justify">There are many possible applications for our [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''beads], as it is possible to display all kinds of different proteins on their surface. To easily create them, we designed a [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks/Sporovector '''Sporo'''vector] in which you just have to insert the gene encoding your fusion protein of choice. Since there are so many possible applications, we illustrate three examplary ideas for future '''Sporo'''beads in the following section:</p> | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| | <div class="box"> | | <div class="box"> |
| | ====Project Navigation==== | | ====Project Navigation==== |


 "
"