Team:LMU-Munich/Application
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| Line 35: | Line 35: | ||
==Protein Screening== | ==Protein Screening== | ||
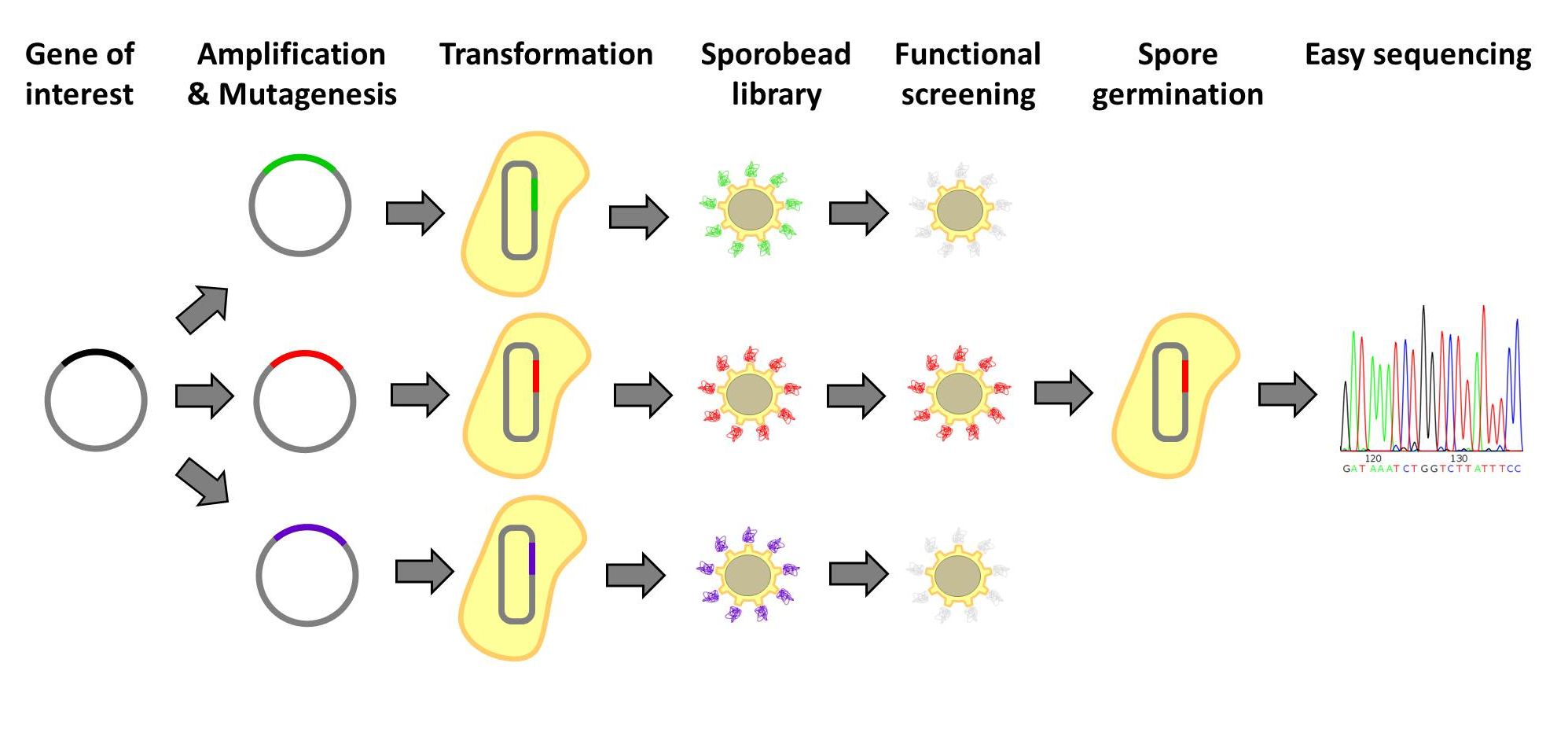
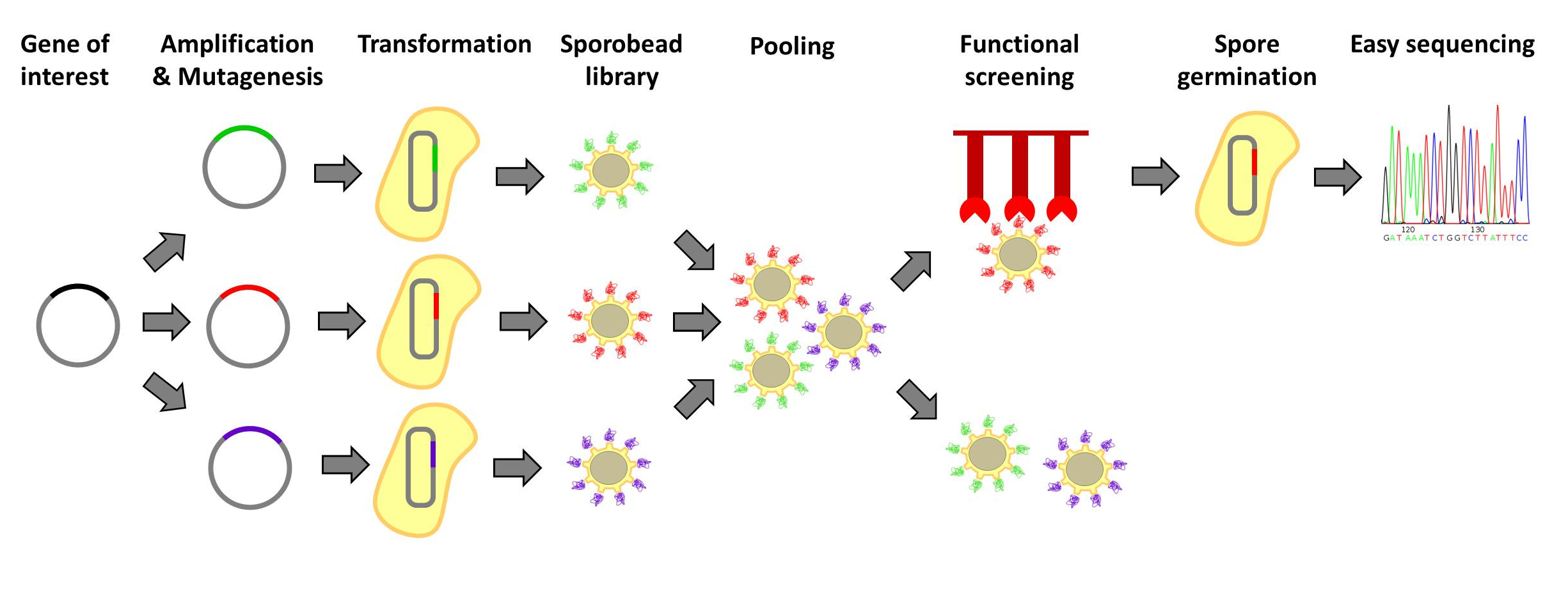
| - | <p align="justify">Designer protein molecules, which bind their desired targets specifically, have numerous uses. Designer proteins are frequently created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in this method. Despite the success of the method, it is labor intensive and limited in throughput. Individual mutated proteins must be tracked throughout the entire screening process. Our '''Sporo'''beads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. | + | <p align="justify">Designer protein molecules, which bind their desired targets specifically, have numerous uses. Designer proteins are frequently created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in this method. Despite the success of the method, it is labor intensive and limited in throughput. Individual mutated proteins must be tracked throughout the entire screening process. Our '''Sporo'''beads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. Figures 1 and 2 offer schematics of the process of using '''Sporo'''beads for protein screening for tests and affinities.</p> |
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {|align:center | ||
| + | |[[File:protein_libraries.jpg|620px|center]] | ||
| + | |- | ||
| + | | style="width: 80%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Fig. 1: '''The simple protein-screening process in our Sporobeads'''. </font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {|align:center | ||
| + | |[[File:protein_libraries_affinities.jpg|620px|center]] | ||
| + | |- | ||
| + | | style="width: 80%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Fig. 1: '''The simple protein-screening process in our Sporobeads''' using affinity binding as a requisite. </font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
==Further Applications== | ==Further Applications== | ||
Revision as of 13:10, 25 October 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


What are the potential advantages of using B. subtilis spores as functional bioparticals?
They are:
| Very stable even under severe environmental conditions | |
| Easy and cheap to produce in large quantities (check out the link to this commercial offer) | |
| Part of human nutrition in terms of food supplements and generally regarded as safe | |
| Moreover, our Sporobeads are: | |
| Unable to proliferate and therefore most likely not treated as genetically modified organisms by the law | |
Our Sporobeads offer a wide variety of applications within the laboratory, and around the world. Sporobeads can be used for filtration and protein screening.
Filtration
Our Sporobeads can express proteins on their outer coats to bind specific molecular targets. Such targets include heavy metals, toxins, and plastic.
One example of such a filtering protein could be a CPX-Sporobead. CPX is a peptide developed by the 2007 MIT iGEM team, which is extremely hydrophilic, and thus capable of binding microparticles of polystyrene from water. The excessive use of disposable plastic and the lack of universal recycling programs has led to the pollution of the world's oceans. In the ocean, large pieces of plastic litter are ground by sea currents and degraded by UV radiation into microscopic pieces, so called "plastic plankton," which is consumed by fish, filter feeders, and other marine organisms. Such plastic uptake can lead to poisoning, sterility and death.
CPX-Sporobeads in huge filter boxes could be put into place to mechanically filter microscopic plastic particles out of the water. Such specific filtration would be superior to blanket filtration systems, which also remove living phytoplankton important to ocean ecosystems. To prevent the beads from being released into the sea and to ensure the plastic be removed from the water, the Sporobeads could be attached to membranes in the filter boxes. Then the Sporobeads would need to not only display CPX but also a membrane binding protein on their surface.
Protein Screening
Designer protein molecules, which bind their desired targets specifically, have numerous uses. Designer proteins are frequently created by constructing and screening libraries of mutated protein variants. Proteins with desired properties are selected for in this method. Despite the success of the method, it is labor intensive and limited in throughput. Individual mutated proteins must be tracked throughout the entire screening process. Our Sporobeads would eliminate the need to track protein mutants, allowing researchers to screen huge quantities of mutated proteins, only identifying and sequencing these proteins after successful screening. Figures 1 and 2 offer schematics of the process of using Sporobeads for protein screening for tests and affinities.
|
|
Further Applications
Ultimately, the ability to express virtually any protein of interest shows great potential for laboratory work. Our Sporobeads could additionally express TAL effectors, for the binding of sequence-specific DNA stretches. This could allow simple GMO detection of food crops. Besides simply expressing proteins capable of binding elements of interest, our Sporobeads could express proteins which have enzymatic activity. The 2011 University of Washington iGEM team developed an enzyme, Kumamolisin, which cleaves peptides. The specific substrate of this enzyme is the specific amino acid sequence which causes reactions in individuals with Celiac disease. Such a cleaving enzyme could be used to eliminate irritants from gluten-containing food products.
There are many possible applications for our Sporobeads, as it is possible to display all kinds of different proteins on their surface. To easily create them, we designed a Sporovector in which you just have to insert the gene encoding your fusion protein of choice. Since there are so many possible applications, we illustrate three examplary ideas for future Sporobeads in the following section:
Kumamolisin-Sporobeads
the solution for carefree enjoyment of everyday meals for Celiac sufferers!
World-wide, one out of 3350 people cannot eat meals that contain wheat products or other foods with traces of gluten. This disease is known as Celiac disease. Kumamolisin is an enzyme that cleaves peptides and was produced by the iGEM-Team from the University of Washington last year. The substrate includes a specific sequence of amino acids, which causes Celiac disease in sensitive people when they consume food containing gluten. Our beads could carry Kumamolisin as a secure vehicle for the passage to the stomach, so that the enzyme can work properly where it is needed. The GerminationSTOP we integrated in our spores would prevent outgrowth and ensure a correct dosage. This project is a pharmaceutical application and therefore would have to fulfill the legal requirements for pharmaceuticals. This includes several verification steps of non-toxicity and efficacy (for more information see release for application.
CPX-Sporobeads
rescuing the ocean from human debris!
The excessive use of disposable plastic and the lack of universal recycling programs has led to the pollution of the world's oceans. In the ocean, large pieces of Polystyrene litter are ground by sea currents and degraded by UV radiation into very small pieces, so called "plastic plankton," which is consumed by fish, filter feeders, and other marine organisms. Such plastic uptake can lead to poisoning, sterility and death. The CPX-peptide, created by the 2007 MIT iGEM team, can bind to Polystyrene. CPX-Sporobeads in huge filter boxes could be put into place to mechanically filter microscopic plastic particles, like Polystyrene microparticles, out of the water. Such specific filtration would be superior to blanket filtration systems, which also remove living phytoplankton important to ocean ecosystems. To prevent the beads from being released into the sea and to ensure the plastic be removed from the water, the Sporobeads could be attached to membranes in the filter boxes. Then the Sporobeads would need to not only display CPX but also a membrane binding protein on their surface. [for security information see release for application]
TALE-Sporobeads
easy and cheap detection of genetically modified organisms!
Since the 1990s, green biotechnology has released many transgenic plants into the environment by selling genetically modified seeds. Thus, organic farmers need to prove that their products meet the requirements for organic crops. Usually they pay laboratories to attest the lack of contamination with genetically modified crops. New tools for sequence-specific DNA-binding of molecular biology, TAL effectors, combined with our Sporobeads could be an easy and cheap solution for organic agriculture. Farmers could use a kit with TALE-lacZ-Sporobeads to detect sequences specific for GM-crops. As our Sporobeads are stable and safe vehicles, they could be sent by mail without any extra security precautions. The kit would be suitable for use outside of laboratory. The protocol for this could work as follows:
DNA extracted from plants with solutions provided by the kit is immobilized and fixed on a nitrocellulose membrane. This membrane is then washed, incubated with Sporobeads in solution and washed again. With addition of a substrate, the surface displayed LacZ of bound Sporobeads will catalyze a reaction so that a blue staining appears. If no such DNA is present, the spores will not bind and no blue color will appear.
 "
"