Team:LMU-Munich
From 2012.igem.org
| (25 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
| - | [[File:Prezi Home.png|600px]] | + | [[File:Prezi Home.png|600px|link=Team:LMU-Munich/Project]] |
<div class="box"> | <div class="box"> | ||
| - | {| | + | {| |
| - | |[[File: | + | |[[File:NEXT.png|100px|link=Team:LMU-Munich/Abstract]] |
| - | | | + | |'''Bead'''zillus - For a guided tour through the most important information about our project and results, klick [[Team:LMU-Munich/Abstract|here]]. |
|} | |} | ||
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
{| | {| | ||
| - | |[[File: | + | |[[File:LMU_Why_BS.png|100px|link=Team:LMU-Munich/Why_Beadzillus]] |
| - | | | + | |Why is there a need for our Project [[Team:LMU-Munich/Why_Beadzillus|'''Bead'''zillus]]? |
|} | |} | ||
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
{| | {| | ||
| - | |[[File: | + | |[[File:LMU_The_solution.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] |
| - | | | + | |Why we choose to work with [[Team:LMU-Munich/Bacillus_Introduction|''Bacillus subtilis'']]. |
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | {| | ||
| + | |[[File:Requirements.png|95px|link=Team:LMU-Munich/Bacillus_BioBricks]] | ||
| + | |To carry out our project we first to had to create a [[Team:LMU-Munich/Bacillus_BioBricks|<b><i>Bacillus</i>B</b>io<b>B</b>rick<b>B</b>ox]] and had to think over important [[Team:LMU-Munich/Germination_Stop|security issues]]. | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | {| | ||
| + | |[[File:4-Sporobeads.png|95px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | |Finally, we succeeded in displaying proteins on endospores. Our [[Team:LMU-Munich/Spore_Coat_Proteins|'''Sporo'''beads]]. | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="box"> | ||
| + | {| | ||
| + | |[[File:LMU 5-Application.png|90px|link=Team:LMU-Munich/Application]] | ||
| + | |With our '''Sporo'''beads a lot of [[Team:LMU-Munich/Application|applications]] are imaginable. | ||
|} | |} | ||
</div> | </div> | ||
| + | |||
| Line 31: | Line 50: | ||
<p align="justify"> | <p align="justify"> | ||
| - | We chose to work with [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction <i>Bacillus subtilis</i>] to set new horizons and offer tools for this model organism to the <i>Escherichia coli</i>-dominated world of iGEM. Therefore, we created a [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks <b><i>Bacillus</i>B</b>io<b>B</b>rick<b>B</b>ox] (<b>B<sup>4</sup></b>) composed of reporter genes, defined promoters, as well as reporter, expression, and empty vectors in BioBrick standard. <i>B. subtilis</i> naturally produces stress resistant endospores which can germinate in response to suitable environmental conditions. To highlight this unique feature using the <b>B<sup>4</sup></b>, we developed [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins <b>Sporo</b>beads]. These are spores displaying fusion proteins on their surface. As a proof of principle, we fused GFP to the outermost layer. Expanding this idea, we designed a [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks/Sporovector <b>Sporo</b>vector] to easily create any <b>Sporo</b>bead imaginable. Because the <b>Sporo</b>beads must be biologically safe and stable vehicles, we [https://2012.igem.org/Team:LMU-Munich/Germination_Stop prevented germination] by knocking out involved genes and developed a [https://2012.igem.org/Team:LMU-Munich/Germination_Stop <b>Suicide</b>switch] turned on in case of germination. With the project <b>Bead</b>zillus, our team demonstrates the powerful nature of <i>B. subtilis</i>. | + | We chose to work with [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction <i>Bacillus subtilis</i>] to set new horizons and offer tools for this model organism to the <i>Escherichia coli</i>-dominated world of iGEM. Therefore, we created a [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks <b><i>Bacillus</i>B</b>io<b>B</b>rick<b>B</b>ox] (<b>B<sup>4</sup></b>) composed of reporter genes, defined promoters, as well as reporter, expression, and empty vectors in BioBrick standard. <i>B. subtilis</i> naturally produces stress-resistant endospores which can germinate in response to suitable environmental conditions. To highlight this unique feature using the <b>B<sup>4</sup></b>, we developed [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins <b>Sporo</b>beads]. These are spores displaying fusion proteins on their surface. As a proof of principle, we fused GFP to the outermost layer. Expanding this idea, we designed a [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks/Sporovector <b>Sporo</b>vector] to easily create any <b>Sporo</b>bead imaginable. Because the <b>Sporo</b>beads must be biologically safe and stable vehicles, we [https://2012.igem.org/Team:LMU-Munich/Germination_Stop prevented germination] by knocking out involved genes and developed a [https://2012.igem.org/Team:LMU-Munich/Germination_Stop <b>Suicide</b>switch] turned on in case of germination. With the project <b>Bead</b>zillus, our team demonstrates the powerful nature of <i>B. subtilis</i>. |
</p> | </p> | ||
| - | |||
| - | |||
| - | |||
Latest revision as of 13:46, 26 October 2012

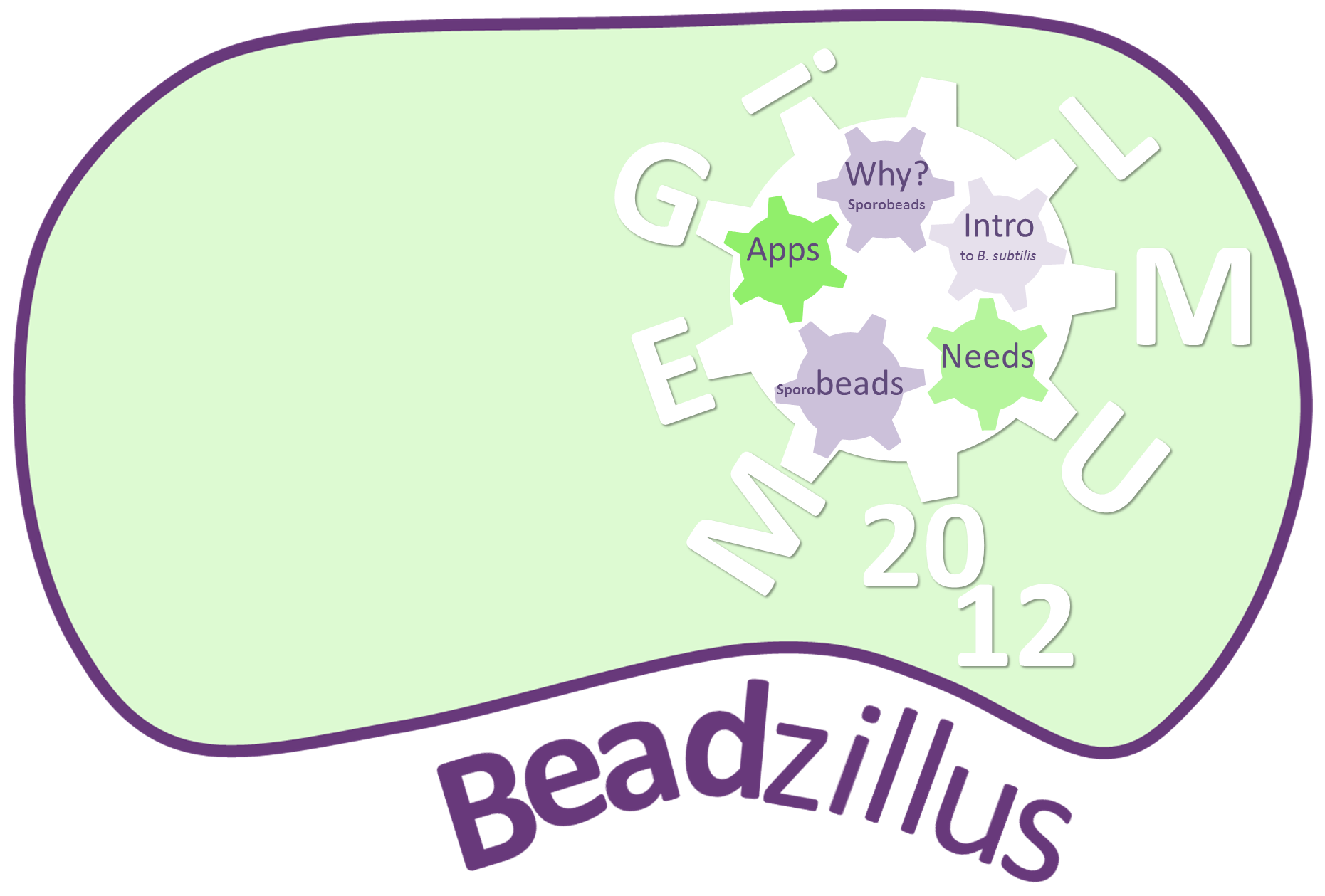
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

| Opening Bacillus subtilis and its unique biology to the iGEM world |
| Beadzillus - For a guided tour through the most important information about our project and results, klick here. |

| Why is there a need for our Project Beadzillus? |

| Why we choose to work with Bacillus subtilis. |

| To carry out our project we first to had to create a BacillusBioBrickBox and had to think over important security issues. |

| Finally, we succeeded in displaying proteins on endospores. Our Sporobeads. |

| With our Sporobeads a lot of applications are imaginable. |
Beadzillus: Fundamental BioBricks for Bacillus subtilis and spores as a platform for protein display
We chose to work with Bacillus subtilis to set new horizons and offer tools for this model organism to the Escherichia coli-dominated world of iGEM. Therefore, we created a BacillusBioBrickBox (B4) composed of reporter genes, defined promoters, as well as reporter, expression, and empty vectors in BioBrick standard. B. subtilis naturally produces stress-resistant endospores which can germinate in response to suitable environmental conditions. To highlight this unique feature using the B4, we developed Sporobeads. These are spores displaying fusion proteins on their surface. As a proof of principle, we fused GFP to the outermost layer. Expanding this idea, we designed a Sporovector to easily create any Sporobead imaginable. Because the Sporobeads must be biologically safe and stable vehicles, we prevented germination by knocking out involved genes and developed a Suicideswitch turned on in case of germination. With the project Beadzillus, our team demonstrates the powerful nature of B. subtilis.
Team LMU Munich 2012
We are the iGEM team of the Ludwig-Maximilians-University (LMU) in Munich. The team consists of 10 dedicated members: 7 students and our 3 advisors. Visit our Team page for more Details.

|

|
Email us!
igem@bio.lmu.de
| Find out who we are! |
Our Gold Sponsors
 "
"