Team:Kyoto/Notebook
From 2012.igem.org
(→Florigen) |
|||
| (571 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | = | + | [[Image:Header_Kyoto_not_home.jpg|975px|link=Team:Kyoto]] |
| - | + | {{Kyoto/header}} | |
| - | + | <html> | |
| - | + | <script type="text/javascript"> | |
| - | + | $(function() { | |
| - | + | showTab("kyoto-tab-NoteFFE"); | |
| - | + | }); | |
| - | + | </script> | |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <ul id="kyoto-tabs"> | |
| - | + | <li><html><a href="" onclick="showTab('kyoto-tab-NoteFFE'); return false;"></html> | |
| - | + | [[Image:KyotoTab_FlowerFairyE.png|link=|NoteFFE]]<html></a></html></li> | |
| - | + | <li><html><a href="" onclick="showTab('kyoto-tab-NoteGGA'); return false;"></html> | |
| - | + | [[Image:KyotoTab_Golden.png|link=|NoteGGA]]<html></a></html></li> | |
| - | + | </ul> | |
| - | + | <div id="kyoto-tab-contents"> | |
| - | + | <div id="kyoto-tab-NoteFFE"> | |
| - | + | {{Kyoto/Notebook/FlowerFairyEcoli}} | |
| - | + | </div> | |
| - | + | <div id="kyoto-tab-NoteGGA"> | |
| - | + | {{Kyoto/Notebook/GoldenGate}} | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | {{Kyoto/footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | '' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | { | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 11:27, 9 October 2012
Caution
| We devided our team into 2 groups in order to achieve Flower Fairy E.coli, Florigen and Secretion group. Florigen aims to confirm expression of florigen in E.coli, activation of R9 peptide, and function of FT made by E.coli. Secretion group aims to make a secretion system without cell death and confirm function of tatABCD and the other proteins. |
August 2
Mutation of FT
by Sato
FT gene has two BioBrick restriction enzyme sites, EcoR1 and Pst1 which is next to each other.
So we tried to delete both at once by using two primers with mutation.
| 10xBufer | 2mM dNTPs | primer fwd | primer rev | template | polymerase | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 1.5 | 1.5 | 0.5 | 1 | 35.5 | 50 |
94℃ 2min, (98℃ 10sec, 68℃ 4min)x4cycles, 4℃ Hold
| PCR product | Dpn1 |
|---|---|
| 50 | 2 |
| product | MilliQ | Ligase | T4 Kinase | Total |
|---|---|---|---|---|
| 2 | 7 | 5 | 1 | 15 |
16℃, 1h incubate
| competent cell | DNA |
|---|---|
| 20 | 2 |
Cells were stored on ice for 30min.
After 42℃ 60sec heat shock, cells were stored on ice for 2min.
Then cells were precultureed at 37℃ for 1hr, plated to Kanamycin plate.
August 13
Liquid culture
FT at 37°C, for overnight.
August 14
Miniprep of FT
by Sato, Takeuchi
The concentration was 81.5ng/uL
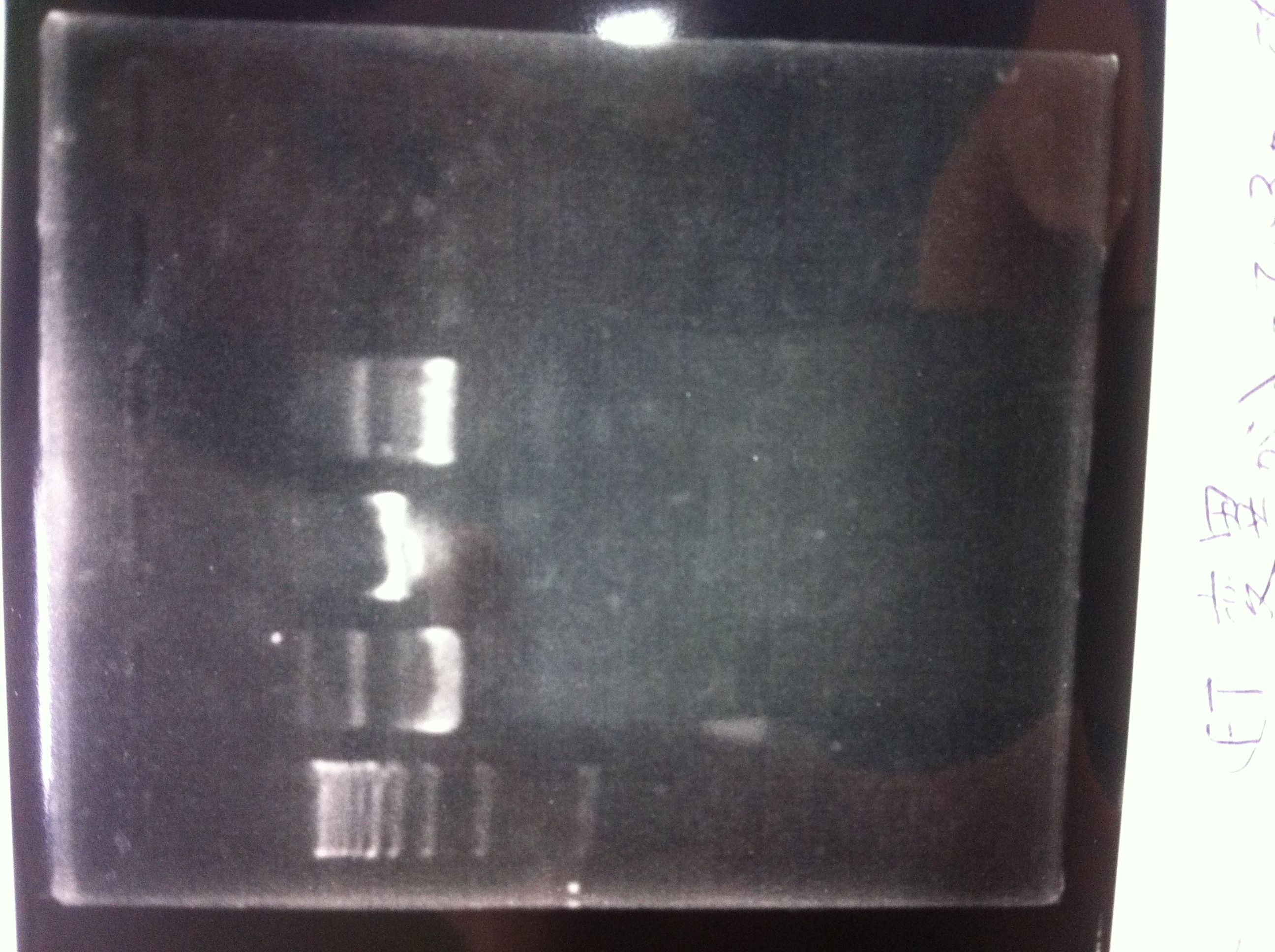
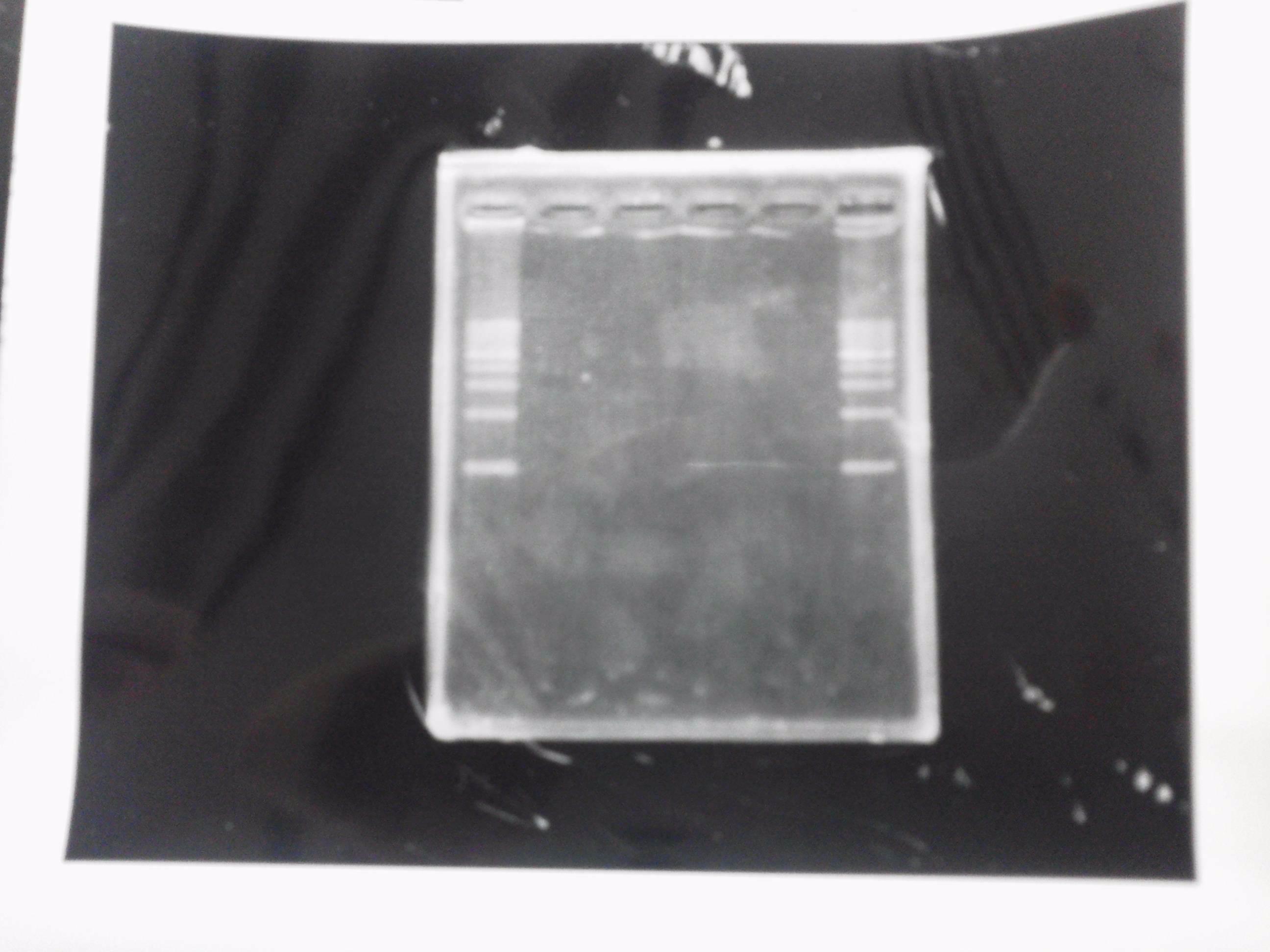
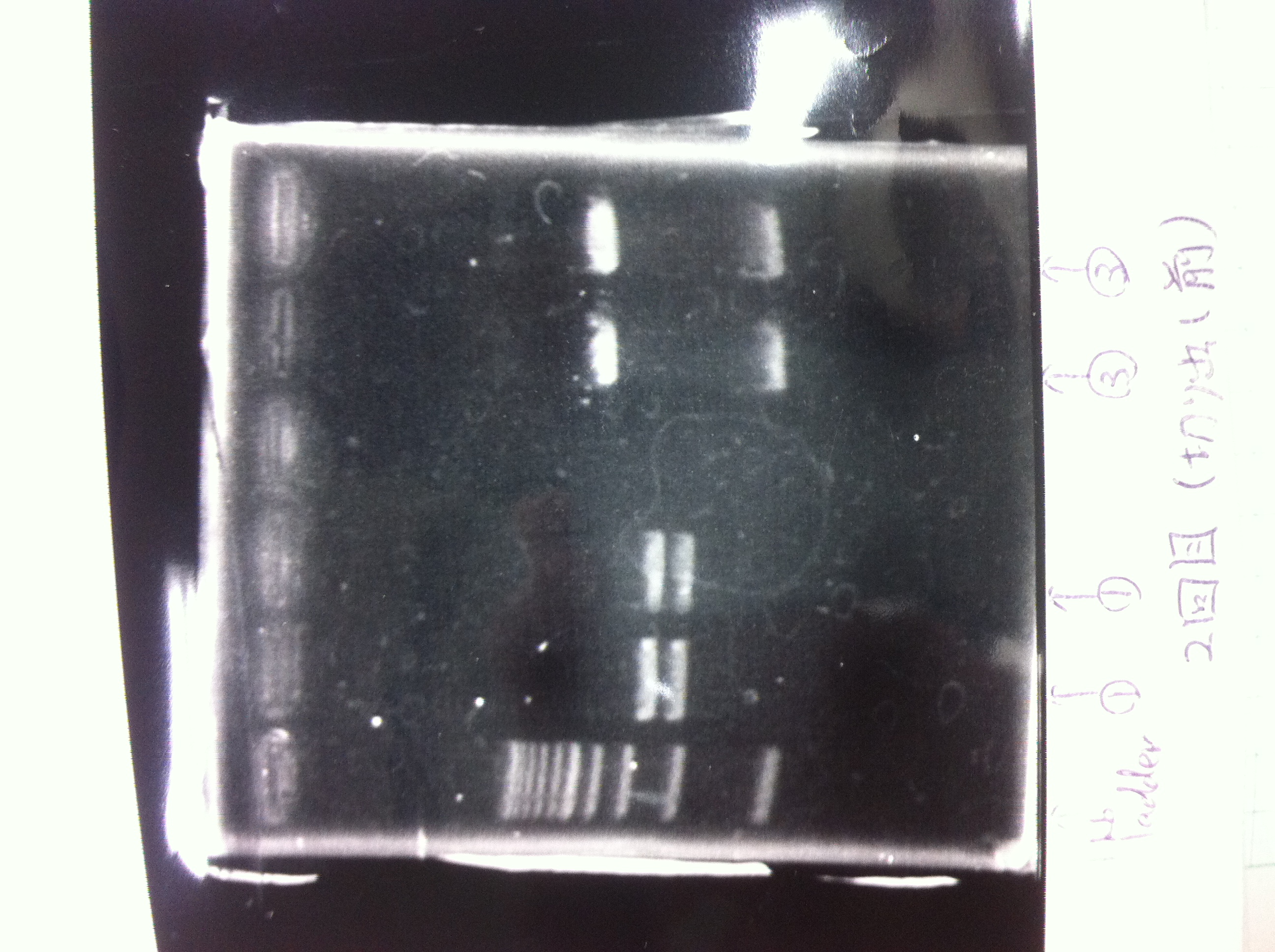
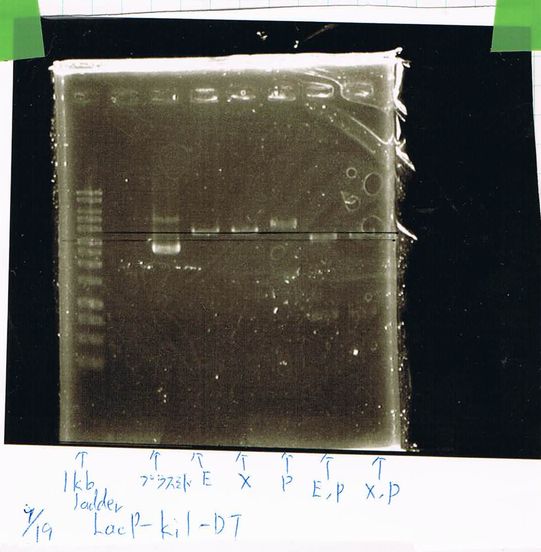
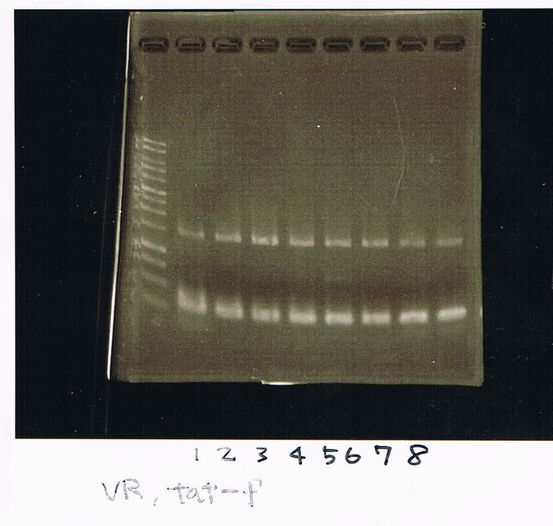
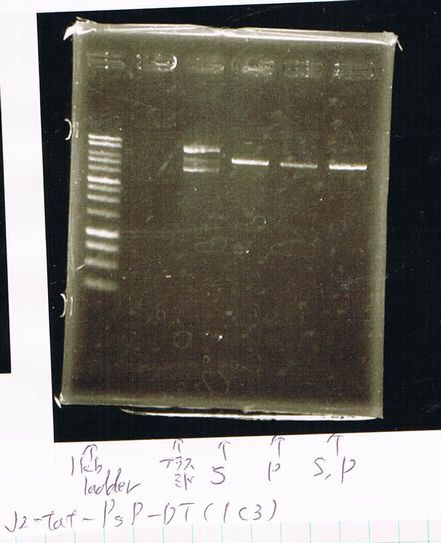
Restriction digestion and Electrophoresis
by Sato
To check wheter mutation was succeed, we did restriction enzyme digestion.
| DNA(FT,80ng/uL) | 10xBuferH | EcoR1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|
| 5 | 2 | 1 | - | 12 | 20 |
| 5 | 2 | - | 1 | 12 | 20 |
37°C 2h incubate
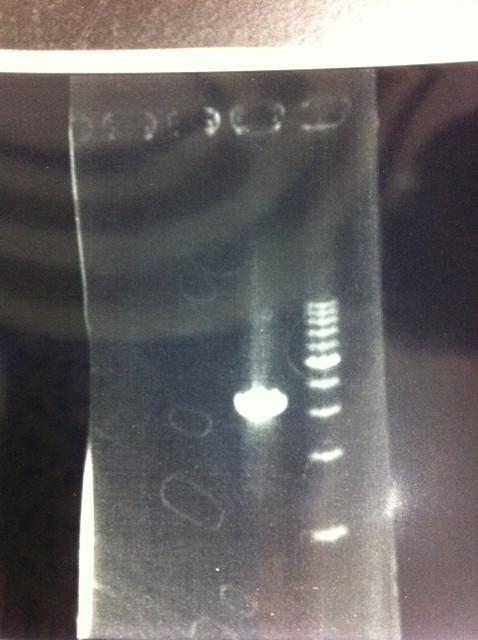
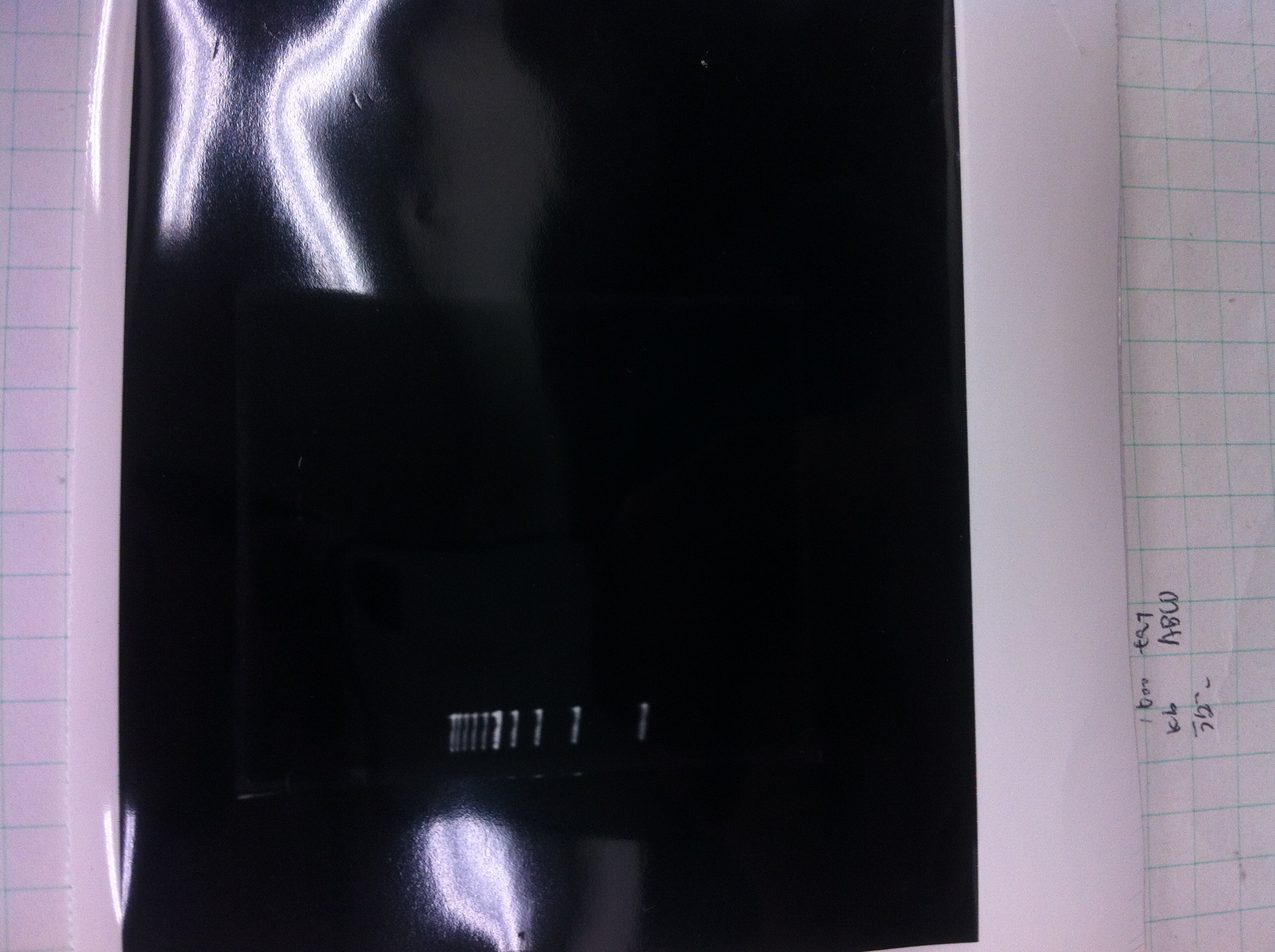
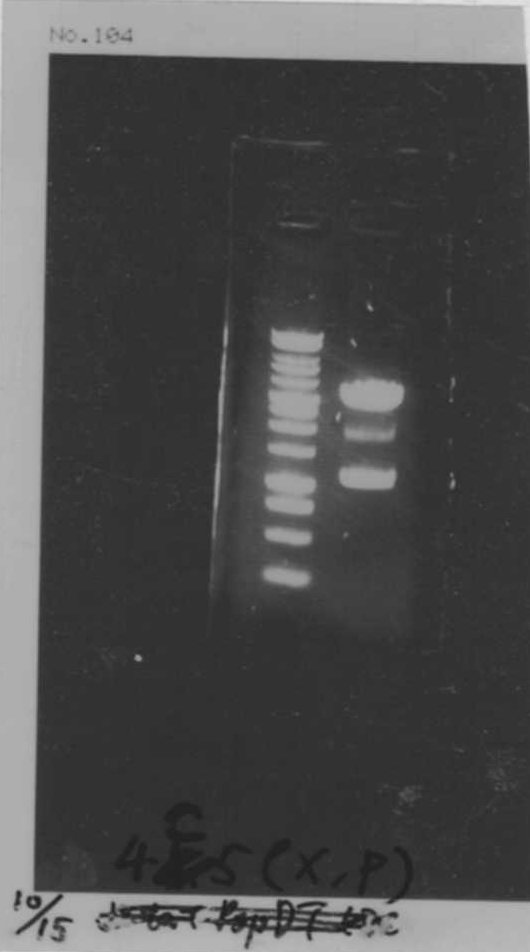
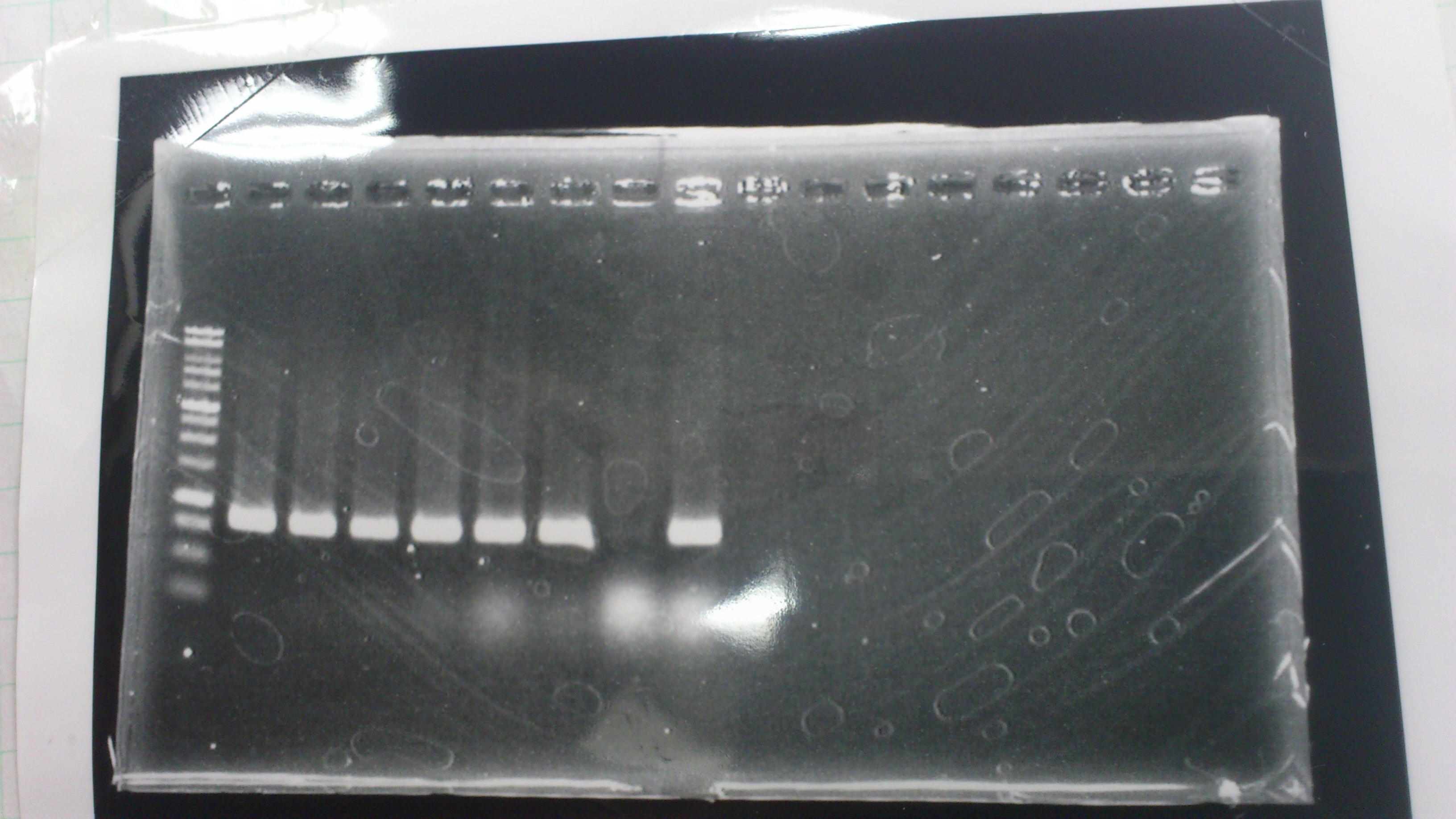
We did electrophoresis of three FT plasmids, non-cutted, EcoR1 cutted, Pst1 cutted.
However, we couldn't get any bands (data not shown.)
Liquid culture
FT (4mL)
August 15
Miniprep of FT
by Sato, Takeuchi, Hyungcheol
We couldn't get enough concentration of plasmids.
Electrophoresis
by Sato
We retried electrophoresis of three samples same as yesterday.
However, we couldn't get any bands as well.
August 16
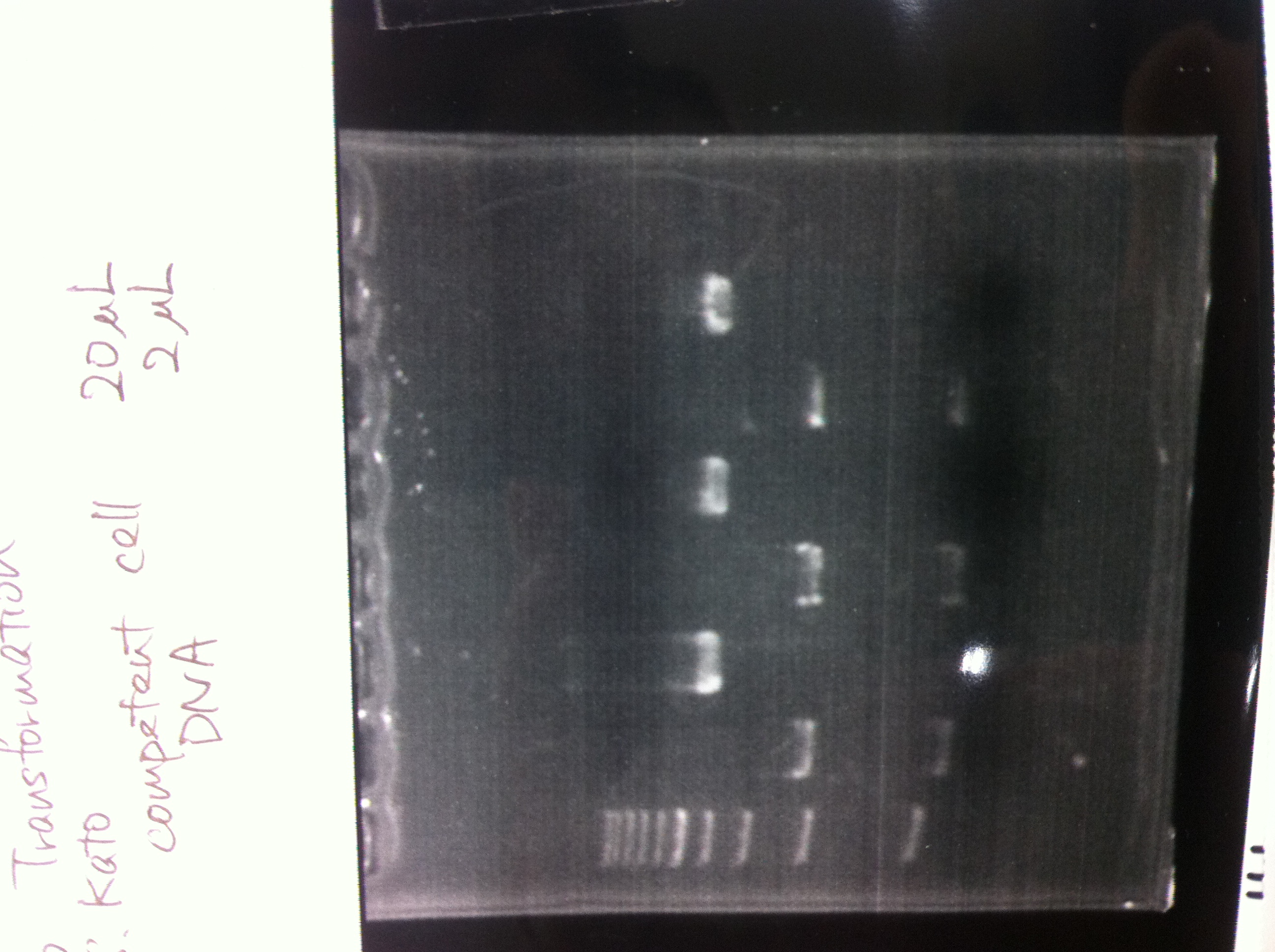
Transformation
by Takeuchi, Ota
| Name | Well | Sample | Competent Cells | Total | Plate | Colony |
|---|---|---|---|---|---|---|
| FT | - | 1 µL | 10 | 11 | LB (Kan+) | × |
| pSB1C3 | 1-3-A | 1 | 10 | 11 | LB (CP+) | ○ |
| I719005 | 1-15-N | 1 | 10 | 11 | LB (Amp+) | ○ |
August 17
Transformation
by Takeuchi
| Name | Well | Sample | Competent Cells | MilliQ | Total | Plate | Colony |
|---|---|---|---|---|---|---|---|
| FT | - | 2 | 2 | 18 | 22 | LB (Kan+) | × |
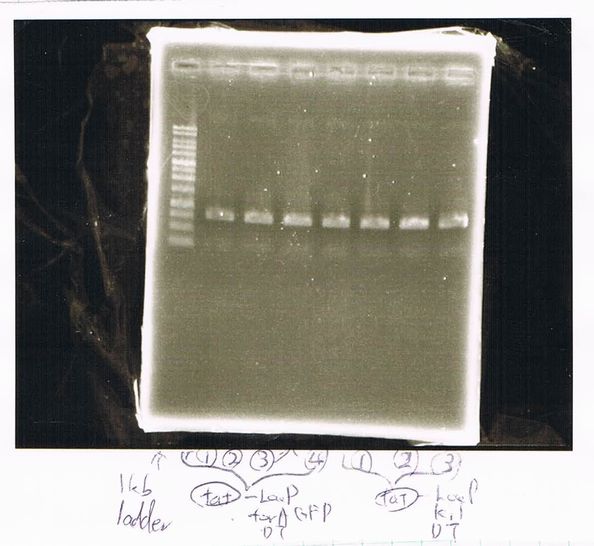
We found that mutation of FT was not successful.
August 20
We decided to do PCR using FT specific primers before mutation.
PCR of FT
by Sato
| 10xBufer | dNTPs | MgSO4 | primer fwd | primer rev | template | polymerase | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1(130ng/µL) | 1(KOD plus neo) | 33 | 50 |
94°C 2min, (98°C 10sec, 68°C 15sec)x30cycles, 4°C Hold
After the ethanol precipitation, we diluted in 30µL of MilliQ.
The concentration was 203ng/µL.
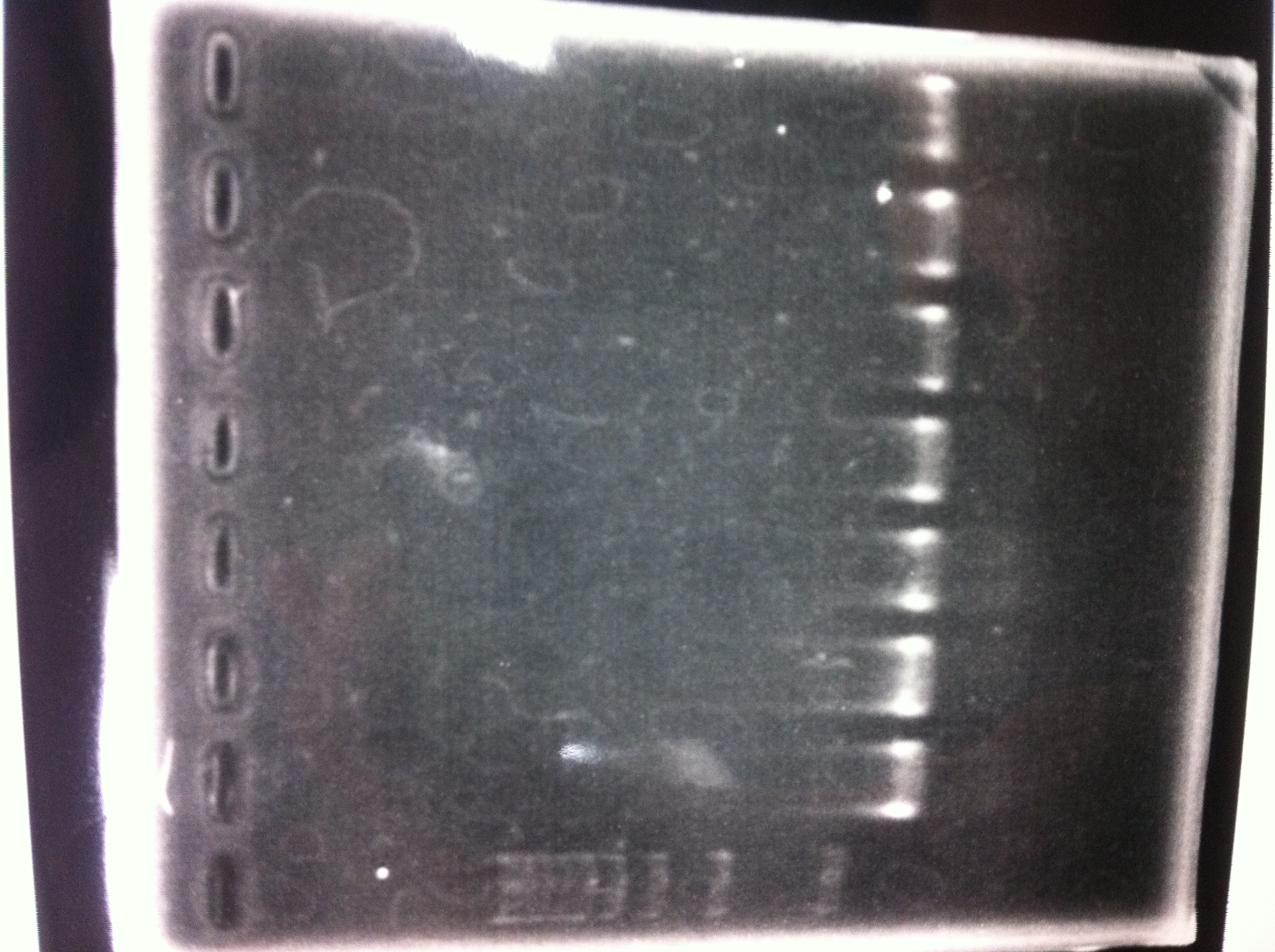
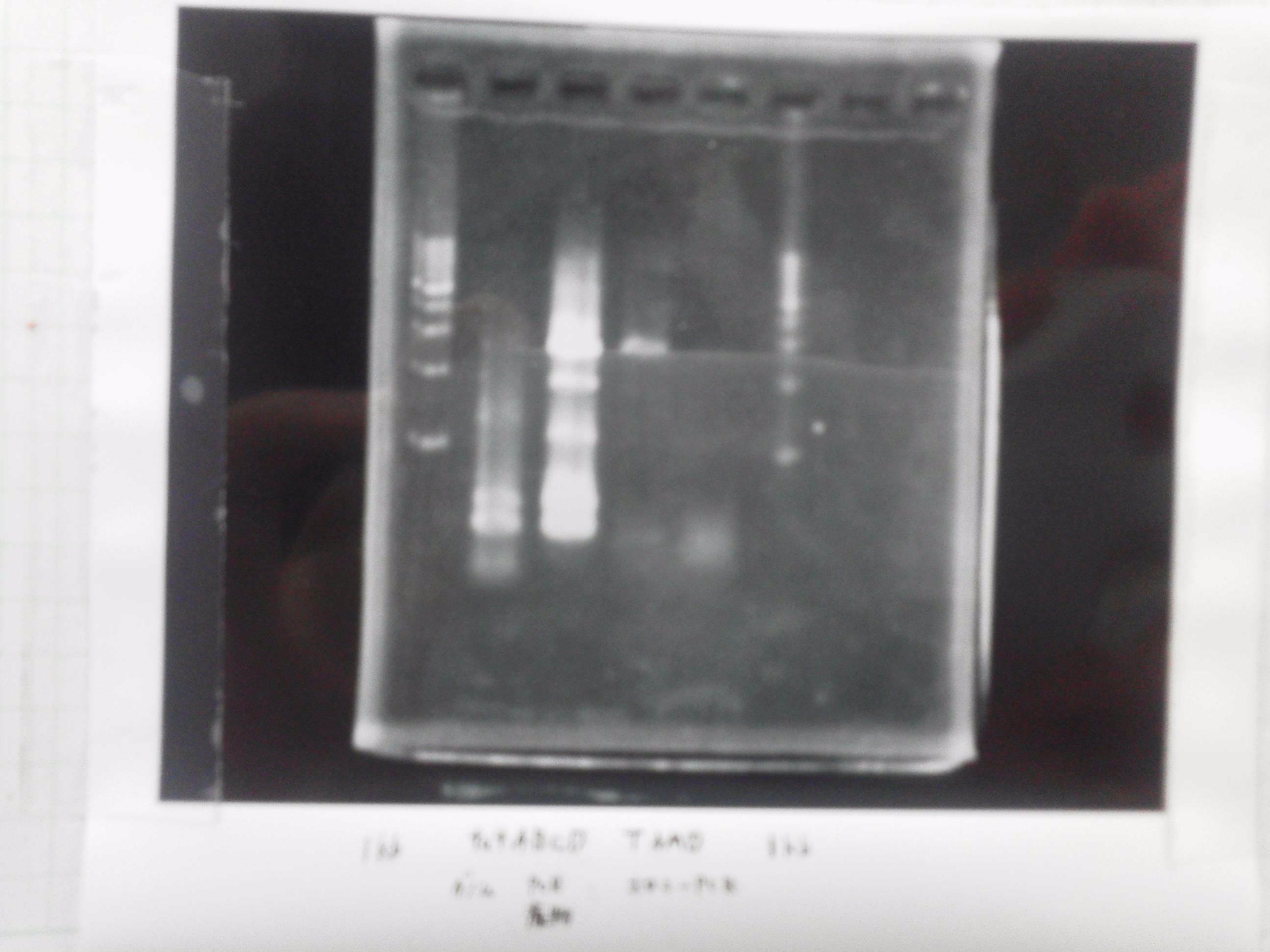
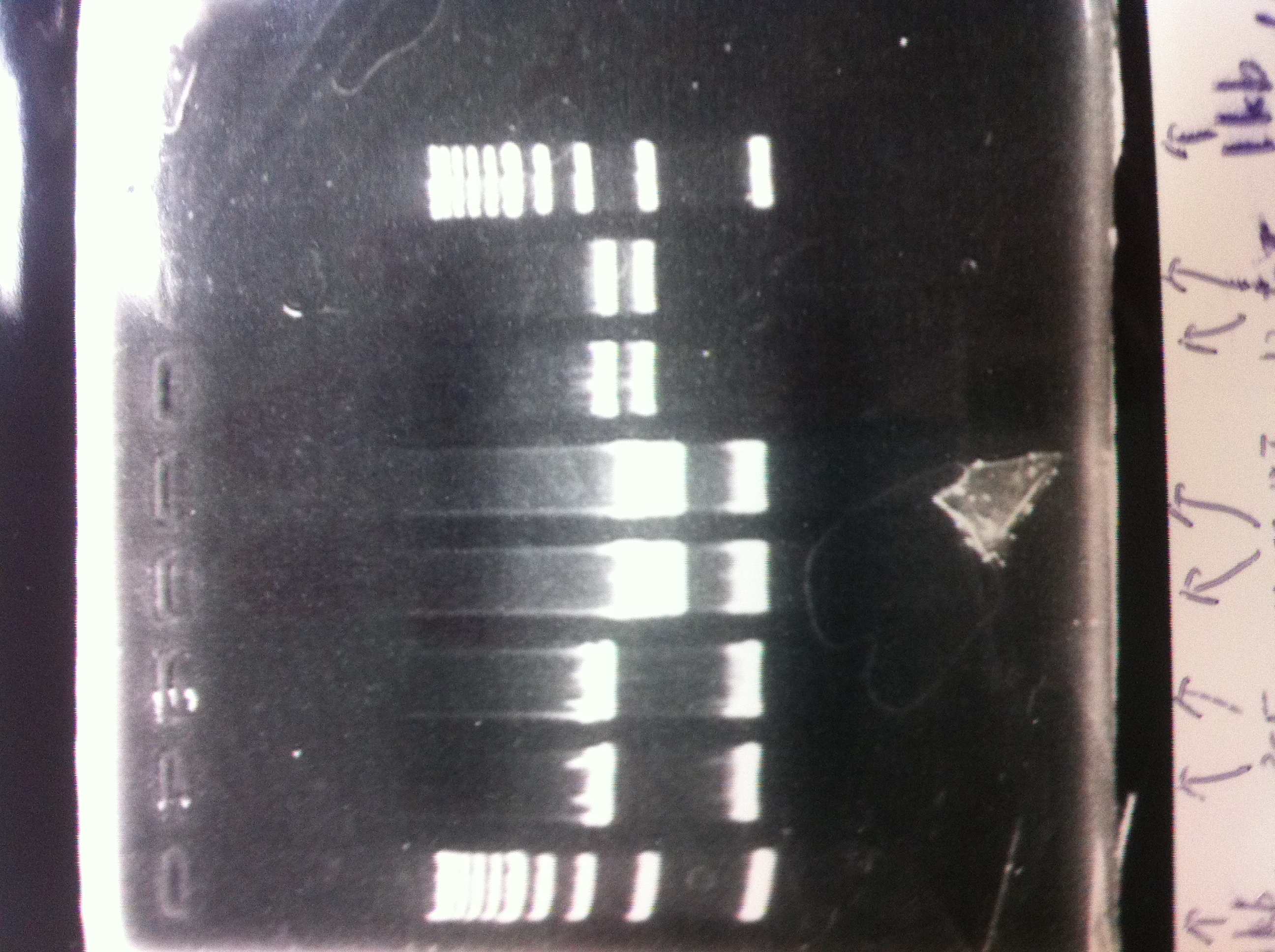
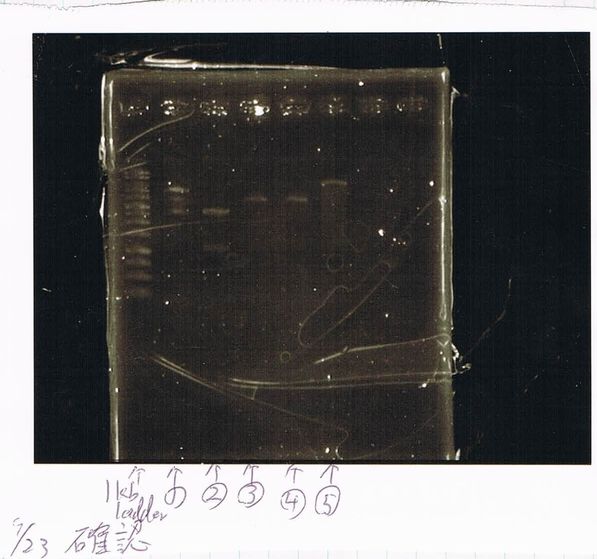
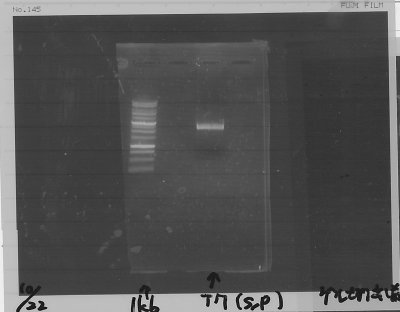
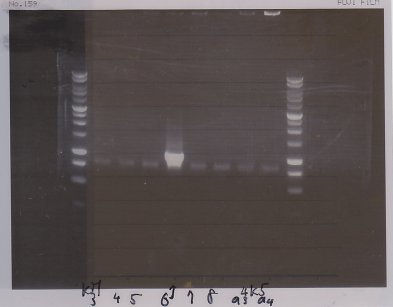
Electrophoresis
| Lane | Name | length(bp) |
|---|---|---|
| 1 | 1kb ladder | - |
| 2 | FT | 600 |
August 21
Restriction digestion
by Sato
| DNA(FT,203ng/µL) | 10xBuferM | Xba11 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 4 | 1 | 1 | 24 | 40 |
37°C, 5h incubate
After the ethanol precipitation, we diluted in 30µL of MilliQ.
The concentration was 54,9ng/µL.
Ligation
by Sato
| Vector | Insert | Ligation High Ver.2 | ||
|---|---|---|---|---|
| pSB1C3 | 1 | FT | 10 | 5.5 |
Liquid culture
T7 promoter, pSB1C3 (4mL)
August 22
Miniprep
by Sato
| T7 promoter | pSB1C3 |
|---|---|
| 85.3ng/µL | 82.93ng/µL |
August 23
Ethanol Precipitation
diluted in 20µL 79.3ng/µL
August 24
Restriction enzyme processing
| T7 promoter(85.3ng/µL) | Spel | Pstl | buffer M | MiliQ | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 2 | 6 | 20 |
->purifying column 33.4ng/µL(dissolution 40µL)
| pSB1C3(82.9ng/µL) | Xbal | Spel | buffer M | MiliQ | Total |
|---|---|---|---|---|---|
| 20 | 1 | 1 | 4 | 14 | 40 |
->gene clean2 39.9ng/µL(dissolution 40µL)
Ligation
| FT(600bp, 79.3ng/µL) | pSB1C3(2000bp,39.9ng/µL) | Ligation High Ver.2 |
|---|---|---|
| 3µL => 597fmol | 2µL => 60fmol | 2.5µL |
| FT(600bp, 79.3ng/µL) | T7(2100bp,33.4ng/µL) | Ligation High Ver.2 |
|---|---|---|
| 2.4µL => 478fmol | 2µL => 48fmol | 2.2µL |
=> 16℃,1hr incubate
August 27
Colony PCR
| 2X Quick Tag | VF2 | VR | MiliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
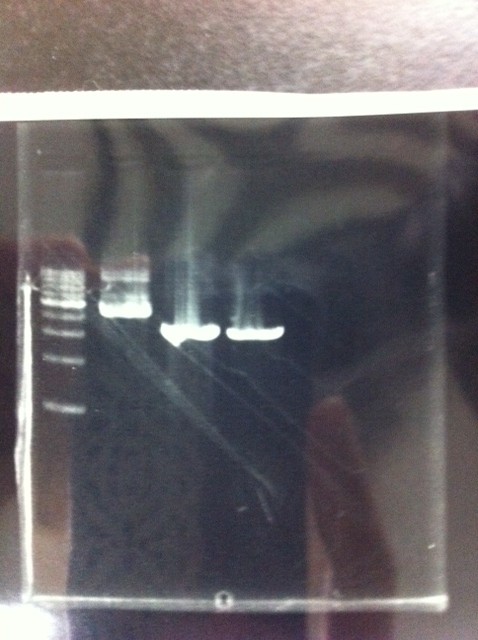
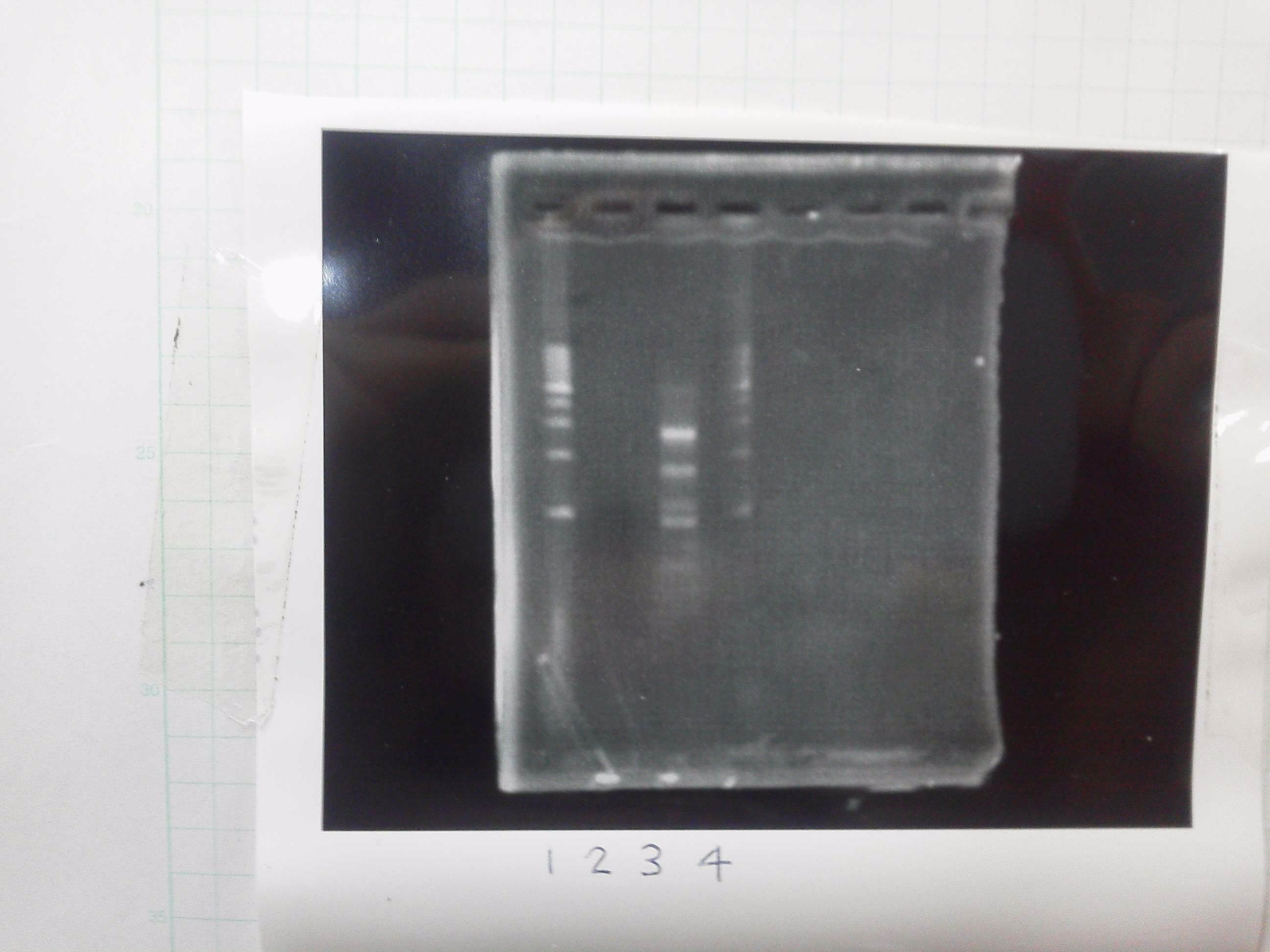
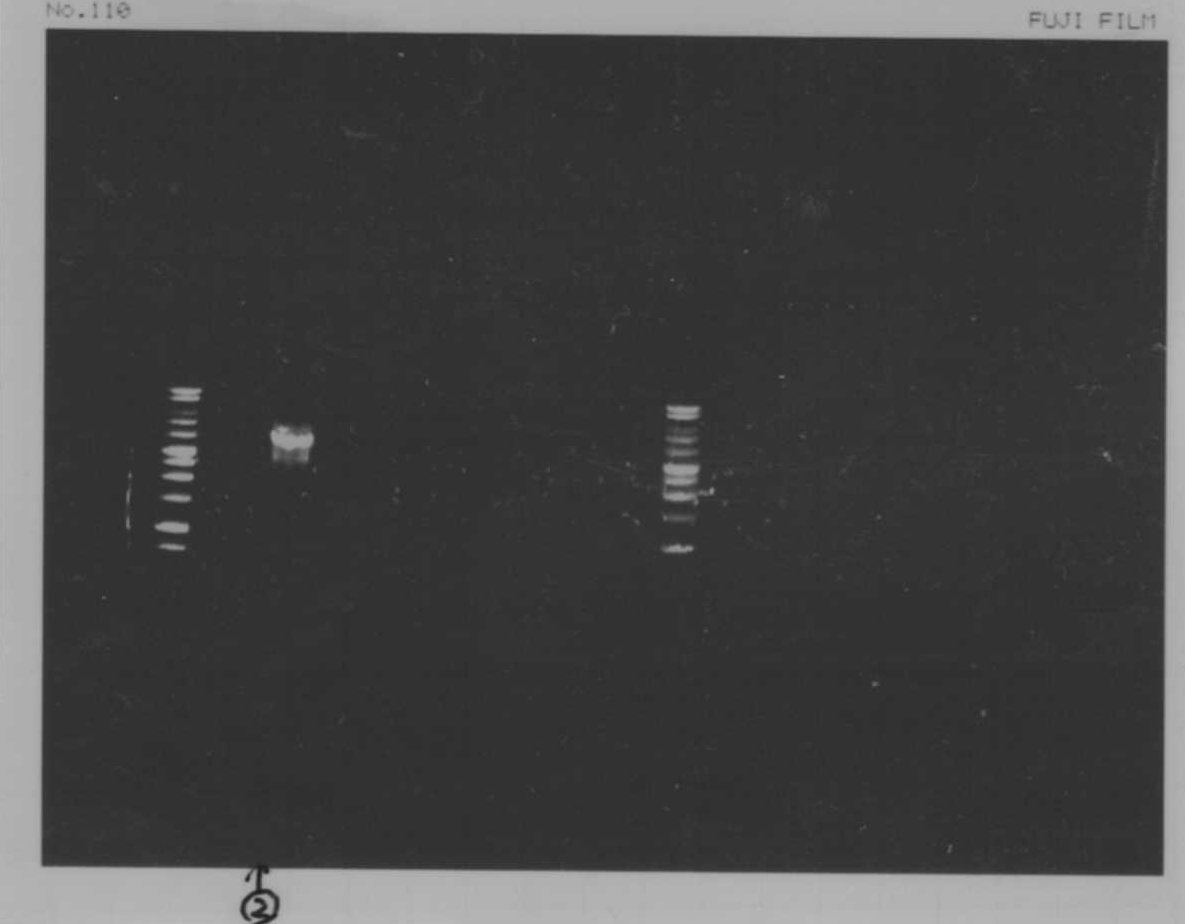
Lane1: 1kb ladder
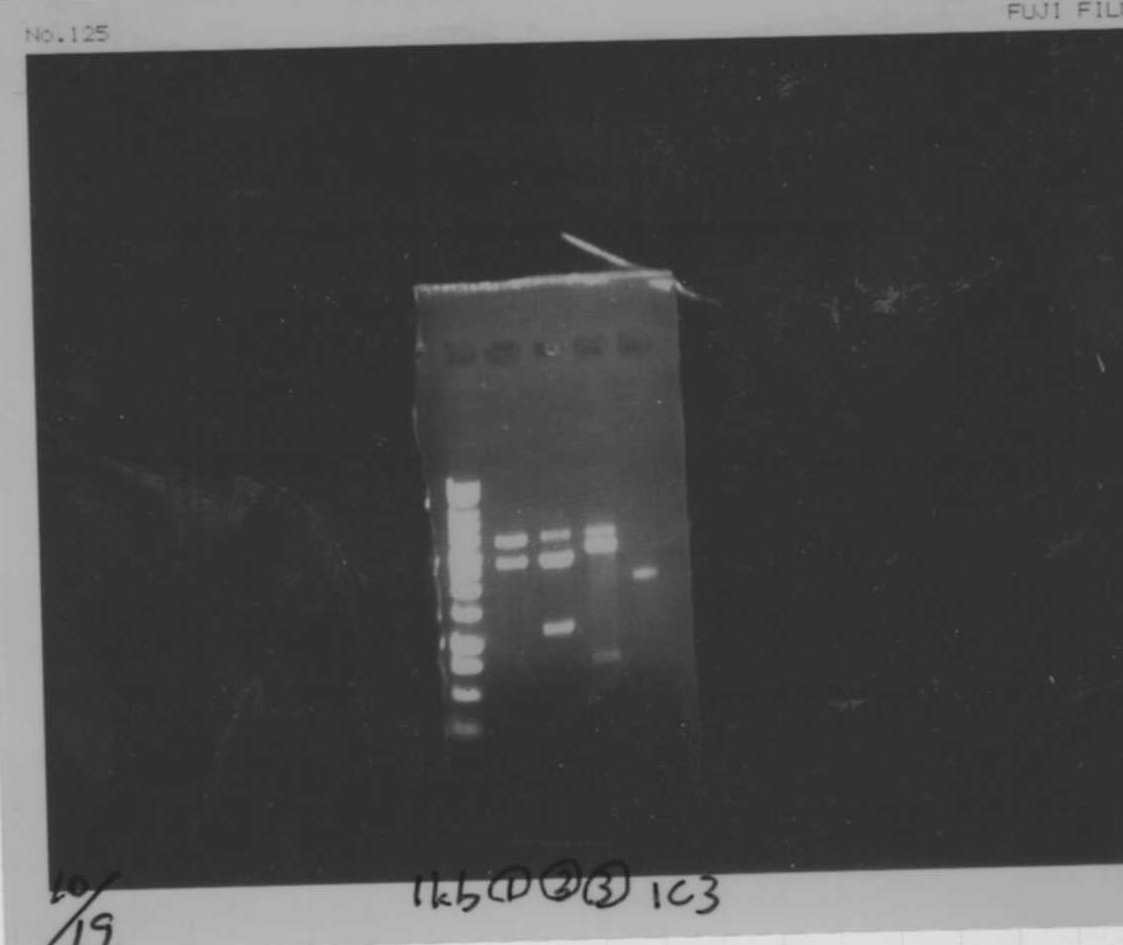
Lane2~16: FT(pSB1C3) about 800bp
FT(TOPO) PCR(re)
| buffer | dNTPs | MgSO4 | primer f | primer r | Template(130ng/µL) | KODplus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 15sec | 30 |
Liquid culture
by Nobeyama
FT 4ml
August 28
Mutation of FT (re)
inverse PCR
| MilliQ | buffer | dNTP | primer f | primer r | FT(130ng/µL) | KODplus | Total |
|---|---|---|---|---|---|---|---|
| 35.5 | 5 | 5 | 1.5 | 1.5 | 0.5 | 1 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 4min | 18 |
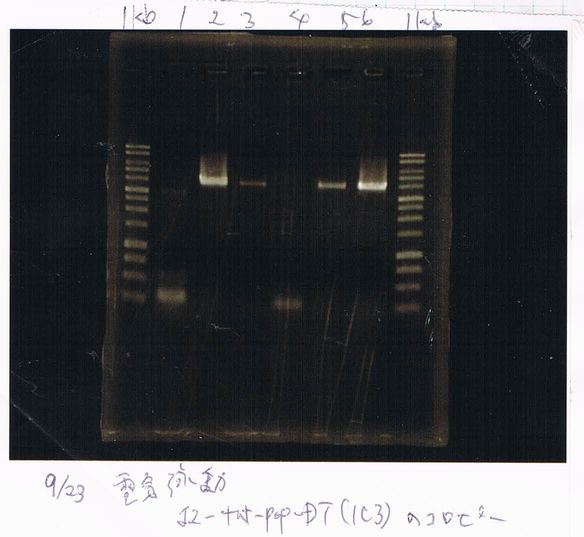
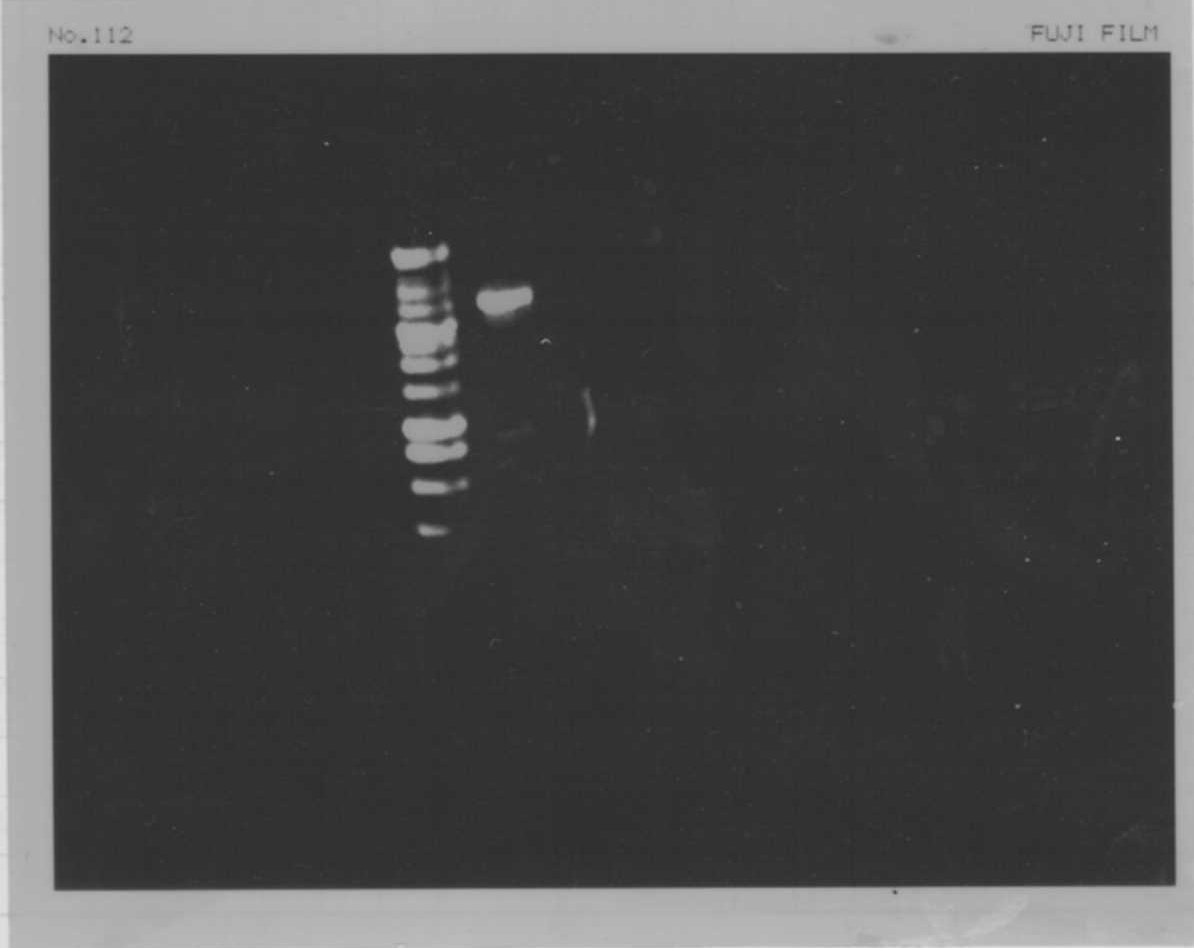
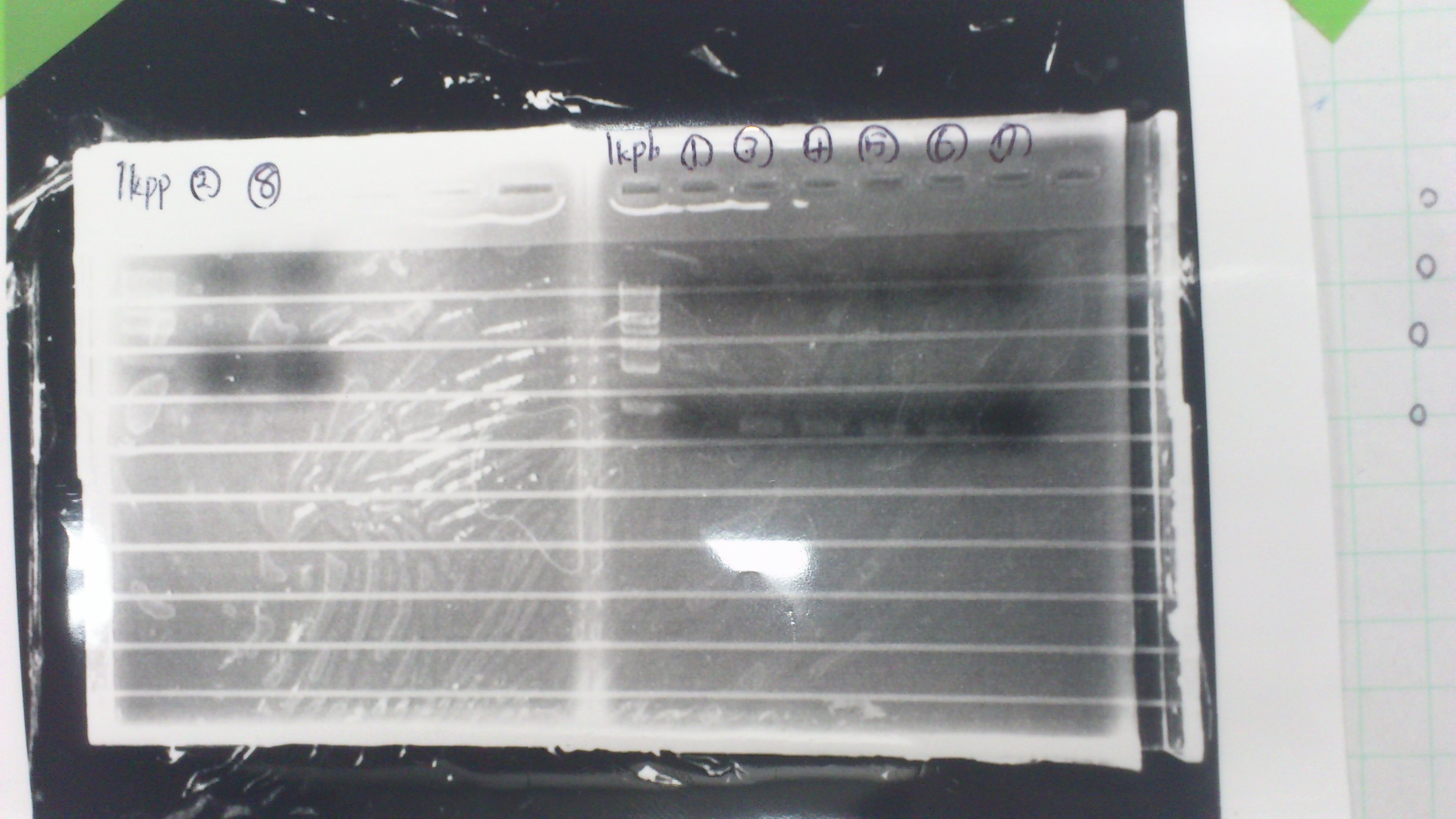
Lane1: 1kb ladder
Lane2: FT
Miniprep FT(TOPO)
158ng/µL
Tranformation
competent cell: 20
BBa.I746902 : 2
(plate 316f)
(GFP generator of pBAD/araC-mut3 GFP:6His-DT)
August 29
Mutaion of FT(re;re)
Inverse PCR
first
| MilliQ | buffer | dNTP | primer f | primer r | FT(130ng/µL) | KODplus | Total |
|---|---|---|---|---|---|---|---|
| 35.5 | 5 | 5 | 1.5 | 1.5 | 0.5 | 1 | 50 |
second
| MilliQ | buffer | dNTP | primer f | primer r | FT(52ng/µL) | KODplus | Total |
|---|---|---|---|---|---|---|---|
| 35 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
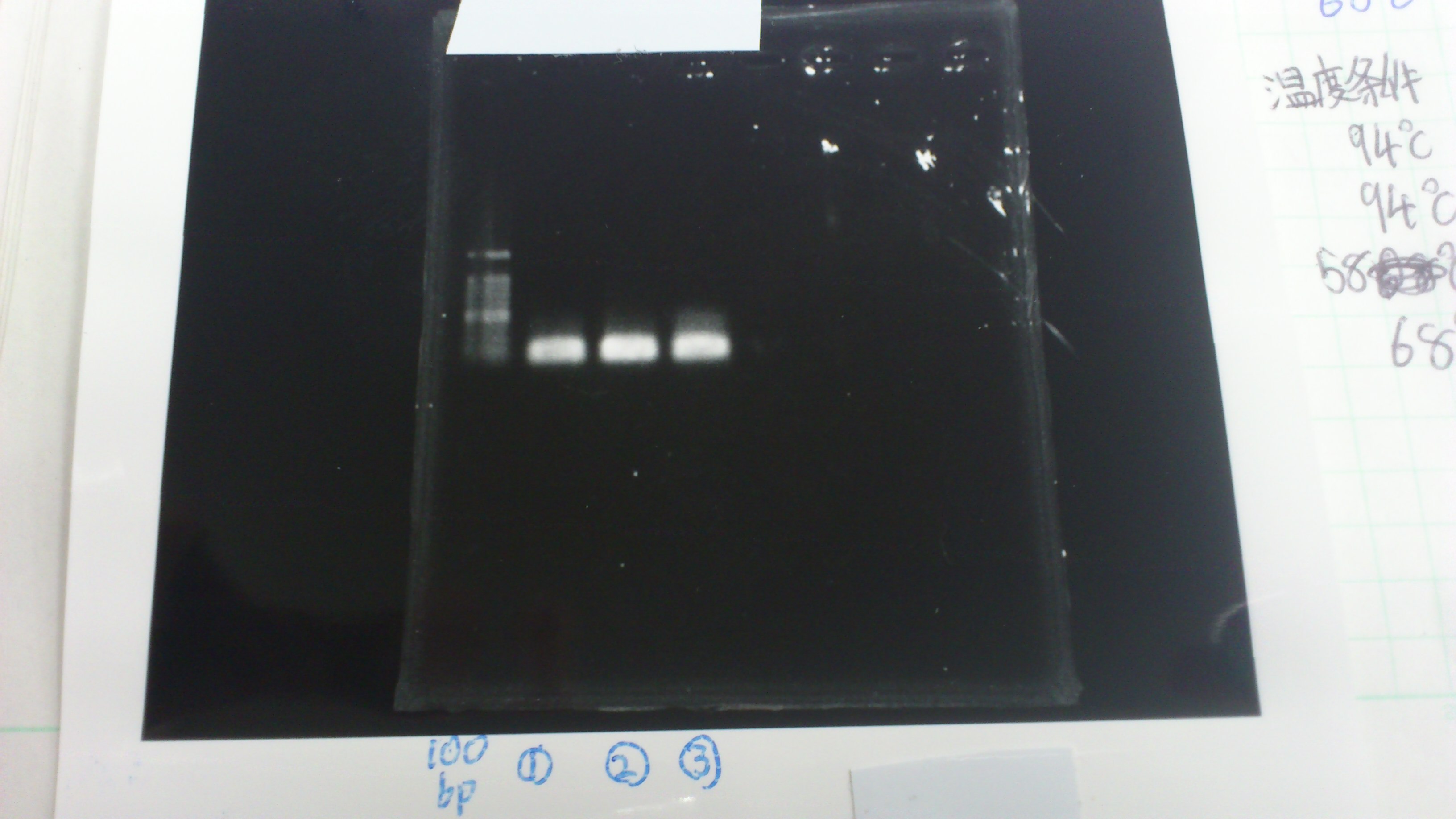
| 2min | 10sec | 4min | 30 |
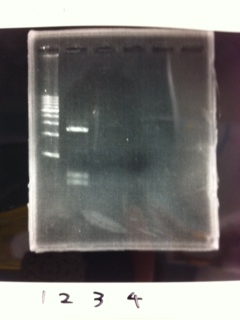
Lane1: 1kb ladder
Lane2: first Template 130ng/µL
Lane3: second Template 53ng/µL
| first sample | Dpnl | total |
|---|---|---|
| 45µL | 2µL | 47µL |
in 37℃, 1 hour
Self-Ligation
| PCR products | MilliQ | Ligation High | T4 kinase | total |
|---|---|---|---|---|
| 2µL | 7µL | 5µL | 1µL | 15µL |
in 16℃, 1.5hour incubate
Transformation
competent cell: 20
DNA : 2
Liquid culture(I746902): 3mL
August 30
Liquid culture(FT) 4mL x2
August 31
Miniprep(FT)
(1) 64.9ng/µL
(2) 52.6ng/µL
Restriction enzyme processing (Mutation checking)
| FT(52.6ng/µL) | bufferH | E.coli | Pst1 | MilliQ | total |
|---|---|---|---|---|---|
| 1µL | 5µL | 0.5µL | 0.5µL | 3.5µL | 10µL |
in 37℃,1.5hour
PCR(RBS primer)
| buffer for KODplus neo | dNTPs | MgSO4 | primer f | primer r | Template(52.6ng/µL) | KODplus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 15sec | 30 |
Lane1: 1kb ladder
Lane2: FT
Lane3: FT(E.coli)
Lane4: FT(Pst1)
Lane5: FT PCR
PCR(re)
| buffer | dNTPs | MgSO4 | primer f | primer r | Template | KOD neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 15sec | 35 |
Lane1: 1kb ladder
Lane2: FT
PCR(re)
| buffer | dNTPs | MgSO4 | primer f | primer r | Template | KOD neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1 | 1 | 1 | 1 | 34 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 10sec | 30 |
September 2
PCR(re;re)
first
| buffer | dNTPs | MgSO4 | primer f | primer r | Template(1ng/µL) | KODplus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
second
| buffer | dNTPs | MgSO4 | primer f | primer r | Template(10ng/µL) | KODplus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 10sec | 25 |
Lane1: first Template 1ng
Lane2: second Template 10ng
Lane3: 1kb ladder
refine first Template => 132ng/µL
September 3
Restriction enzyme processing
| FT(132ng/µL) | bufferM | EcoRI | SpeI | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 2 | 1 | 1 | 6 | 20 |
37℃,overnight => refinement 31.6ng/µL (Elution 40µL)
Ligation
Insert(FT: 31.6ng/µL, 600bp ): 2µL = 26fmol
Vector(DT: 28.0ng/µL, 3300bp): 3µL = 240fmol
Ligation High ver.2 :2.5µL
=> 16℃, 2 hours
Transformation
| competent cell | DNA | plate | colony |
|---|---|---|---|
| 20µL | FT-DT 2µL | Amp | o |
| 20µL | pT7-6His-R9 2µL | Amp | o |
| 10µL | GFP generator 1µL | Amp | o |
September 4
Colony PCR
| Quick Tag | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
| 94℃ | 98℃ | 55℃ | 68℃ | cycles |
|---|---|---|---|---|
| 2min | 30sec | 30sec | 1min | 25 |
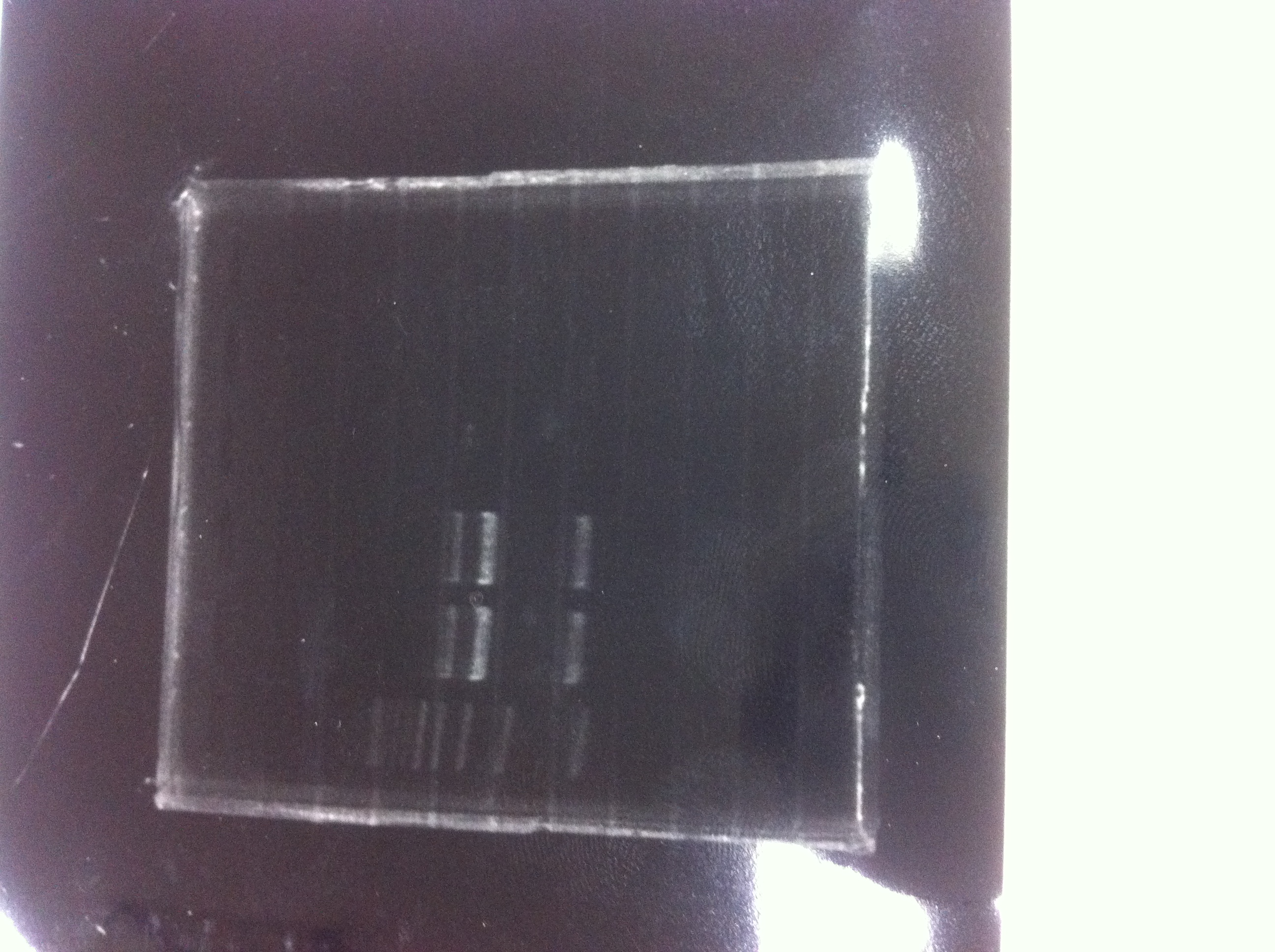
Lane1: 1kb ladder
Lane2: FT-DT(about 700bp)
Liquid culture(4ml) 23:30 ~
GFP generator, FT-DT
September 5
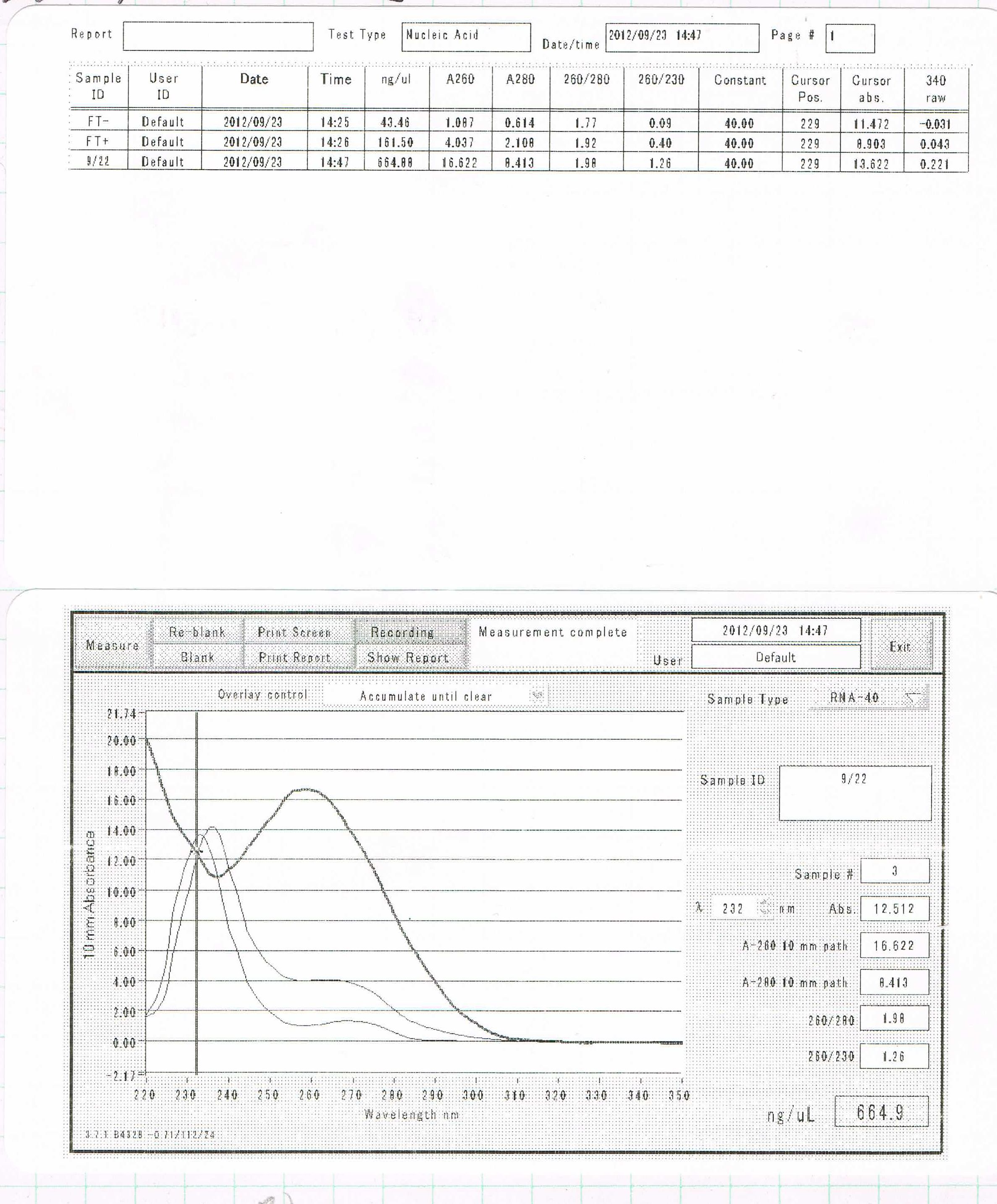
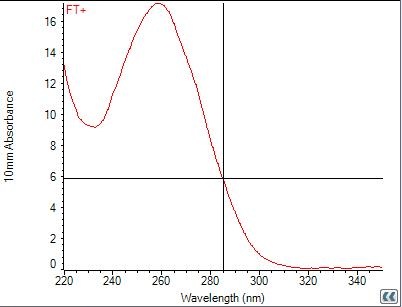
Miniprep(FT-DT) by Sato
88.8ng/µL
Restriction enzyme processing
| FT-DT(88.8ng/µL) | XbaI | PstI | bufferM | MiliiQ | total |
|---|---|---|---|---|---|
| 20 | 1 | 1 | 4 | 14 | 40 |
at 37℃, 2 hours
Electrophoresis by Takeuchi
Restriction enzyme processingby Takeuchi
| FT-DT(88.8ng/µL) | bufferM | XbaI/Spe | MiliiQ | total |
|---|---|---|---|---|
| 4 | 1 | 0.5 | 4.5 | 10 |
| FT-DT | bufferH | Pst/Eco | MiliiQ | total |
|---|---|---|---|---|
| 4 | 1 | 0.5 | 4.5 | 10 |
at 37℃,1hour 10min
September 6
Western blotting(BBa,I746915)
Sample making
SOC(ampt) 50ml + pre culture 1ml x2
OD600 = 0.5~0.7 incubate at 37℃ (OD 0.642)
add IPTG final concentration is 1mM (negative control)
incubate at 37℃,4 hours
SDS-PAGE
Do spin down E.coli and make suspension put E.coli into 1mL 1x sample buffer
95℃,10min
electrophoresis at 500V, 30mA, 50min
Lane1: 10µL (IPTG -)
Lane2: 10µL (IPTG -)
Lane3: 10µL (IPTG +)
Lane4: 5µL (IPTG -)
Lane5: 5µL (IPTG +)
Lane6: 2µL (IPTG -)
Lane7: 2µL (IPTG +)
Blotting at 50V,100mA,30min
Put into blocking buffer and vibrating 30min
Incubate with Anti GFP(1/1000) 10mL at RT,1h
Washing with 10mL TBST (vibrating 10min x2)
Incubate with Anti-mouseAP(1/1000) 10mL at RT, 30min
Washing with 10mL TBST (vibrating 10min x3)
Put NBT,BCIP into dye buffer
September 9
Mutaion of FT
| MilliQ | buffer | dNTP | primer f | primer r | FT(52ng/µL) | KODplus | Total |
|---|---|---|---|---|---|---|---|
| 35 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 4min | 20 |
→ GeneClean II 32.6ng/µL
Dpn1 processing
| TA buffer | DNA | Dpn1 | Total |
|---|---|---|---|
| 3.3 | 31 | 2 | 36.3 |
Ligation
| DNA | MiliiQ | Ligation high Ver.2 | T4 kinase | Total |
|---|---|---|---|---|
| 2 | 7 | 5 | 1 | 15 |
Transformation
competent cell: 10
DNA : 1
September 10
Miniprep of FT
①116.9ng/µL
② 34.5ng/µL
Restriction enzyme processing(Mutation checking)
| FT①/② | bufferHl | EcoRI | PstI | MiliQ | Total |
|---|---|---|---|---|---|
| 4 | 1 | 0.5 | 0 | 4 | 10 |
| 4 | 1 | 0 | 0.5 | 4 | 10 |
PCR
| Bufer | dNTPs | MgSO4 | primer(+/-RBS)fwd | primer(+/-RBS)rev | template①/② | KOD plus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 68℃ | cycles |
|---|---|---|---|
| 2min | 10sec | 10sec | 25 |
->purifying column 17.4ng/µL
Restriction enzyme processing
| FT(RBS+)32.6ng/µL | Xbal | Pstl | buffer M | BSA | Total |
|---|---|---|---|---|---|
| 30 | 1 | 1 | 4 | 4 | 40 |
| T7-His:R9(62.6ng/µL) | SpeI | Pstl | MilliQ | buffer M | Total |
|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 2 | 2 | 20 |
incubate at 37℃, 3hours
->purifying column 17.4ng/µL
September 11
Ligation
Vector(T7: 33.4ng/µL, 2100bp): 1µL = 24fmol
Insert(FT: 17.4ng/µL, 600bp ): 5µL = 217fmol
Ligation High ver.2 : 3µL
=> 16℃, 1 hours
Transformation
| competent cell | DNA |
|---|---|
| 20µL | T7-FT 2µL |
PCR(FT without RBS retry)
| Bufer | dNTPs | MgSO4 | primer(-RBS)fwd | primer(-RBS)rev | template | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1 | 1 | 1 | 1 | 33 | 50 |
| 94℃ | 98℃ | 65℃ | 68℃ | cycles |
|---|---|---|---|---|
| 2min | 15sec | 30sec | 30sec | 25 |
Lane1:FT(RBS-) 600bp
Lane2:Ladder 100bp
=>purifying column 34.6ng/µL
Restriction enzyme processing
| FT(RBS-) | XbaI | PstI | bufferM | BSA | total |
|---|---|---|---|---|---|
| 30 | 1 | 1 | 4 | 4 | 40 |
at 37℃, 2 hours
=> 6.2ng/μL
| GFP-DT | XbaI | PstI | bufferM | BSA | MilliQ | total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 3 | 7 | 30 |
at 37℃, 2 hours
=> 7.0ng/μL
Ligation
Vector(T7-6His-R9: 11.6ng/µL, 2100bp): 1µL = 9fmol
Insert(FT: 6.2ng/µL, 600bp ): 5µL = 79fmol
Ligation High ver.2 : 3µL
=> 16℃, 1 hours
Vector(T7-6His-R9: 11.6ng/µL, 2100bp): 1µL = 9fmol
Insert(GFP-DT: 7.0ng/µL, 1000bp ): 9µL = 95fmol
Ligation High ver.2 : 4µL
=> 16℃, 1 hours
September 12
Transformation
| competent cell | DNA | plate |
|---|---|---|
| 20µL | T7-R9-GFP-DT 2µL | Amp |
| 20µL | T7-R9-FT 2µL | Amp |
| BL21CDE3 10µL | T7-FT 1µL | Amp |
Miniprep (T7-FT)
37.1ng/µL
September 13
Miniprep
①T7-R9-GFP-DT 90ng/µL
②T7-R9-FT 160ng/μL
Restriction enzyme processing
| T7-R9-GFP-DT (90ng/µL) | EcoRI | PstI | buffer H | MilliQ | total |
|---|---|---|---|---|---|
| 20 | 1 | 1 | 3 | 5 | 30 |
at 37℃, 2 hours
=> 20.7ng/μL
| T7-R9-FT (160ng/µL) | EcoRI | SpeI | buffer M | MilliQ | total |
|---|---|---|---|---|---|
| 20 | 1 | 1 | 3 | 5 | 30 |
at 37℃, 2 hours
=> 20.1ng/μL
'Transformation
| competent cell BL21(DE3) | DNA | plate |
|---|---|---|
| 10µL | T7-R9-GFP-DT 1µL | Amp |
| 10µL | T7-R9-FT 1µL | Amp |
Restriction enzyme processing
| Buffer H | BSA | EcoRI | PstI | DpnⅠ | MilliQ | total |
|---|---|---|---|---|---|---|
| 5 | 5 | 0.5 | 0.5 | 0.5 | 13.5 | 25 |
=>We define this solution "2× Master Mix"
| 2× Master Mix | pSB1C3(Linerarized Plasmid Backbone) |
|---|---|
| 4 | 4 |
at 37℃, 30 minutes
80℃, 30 minutes
PCR(Insert His-tag)
| MilliQ | Buffer for iPCR | dNTPs | primer fwd | primer rev | template(T7-FT 37.1ng/μL) | KOD plus | Total |
|---|---|---|---|---|---|---|---|
| 35 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50 |
| 94℃ | 94℃ | 56℃ | 68℃ | cycles |
|---|---|---|---|---|
| 2min | 15sec | 30sec | 2.5min | 15 |
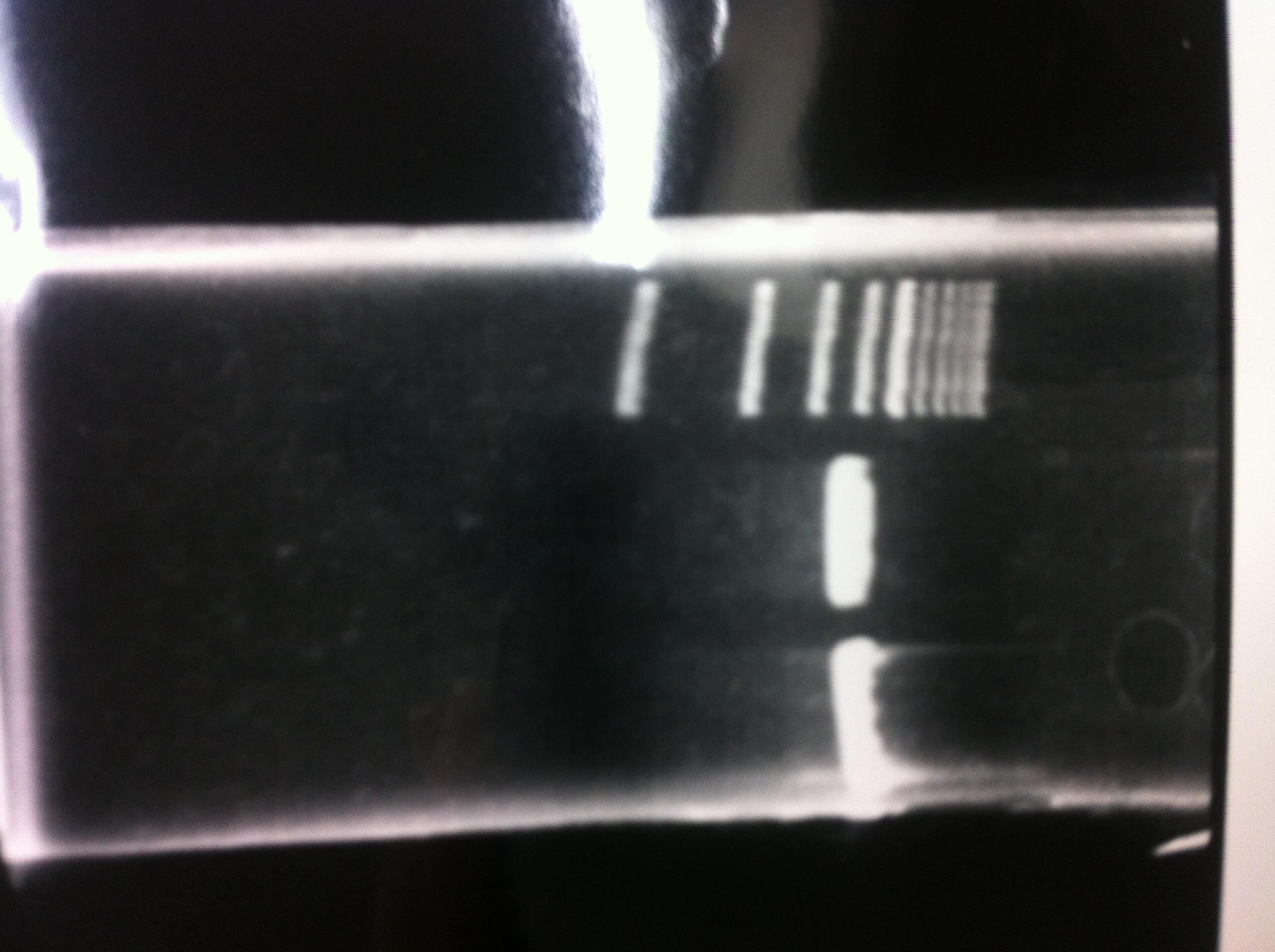
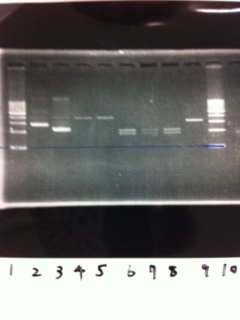
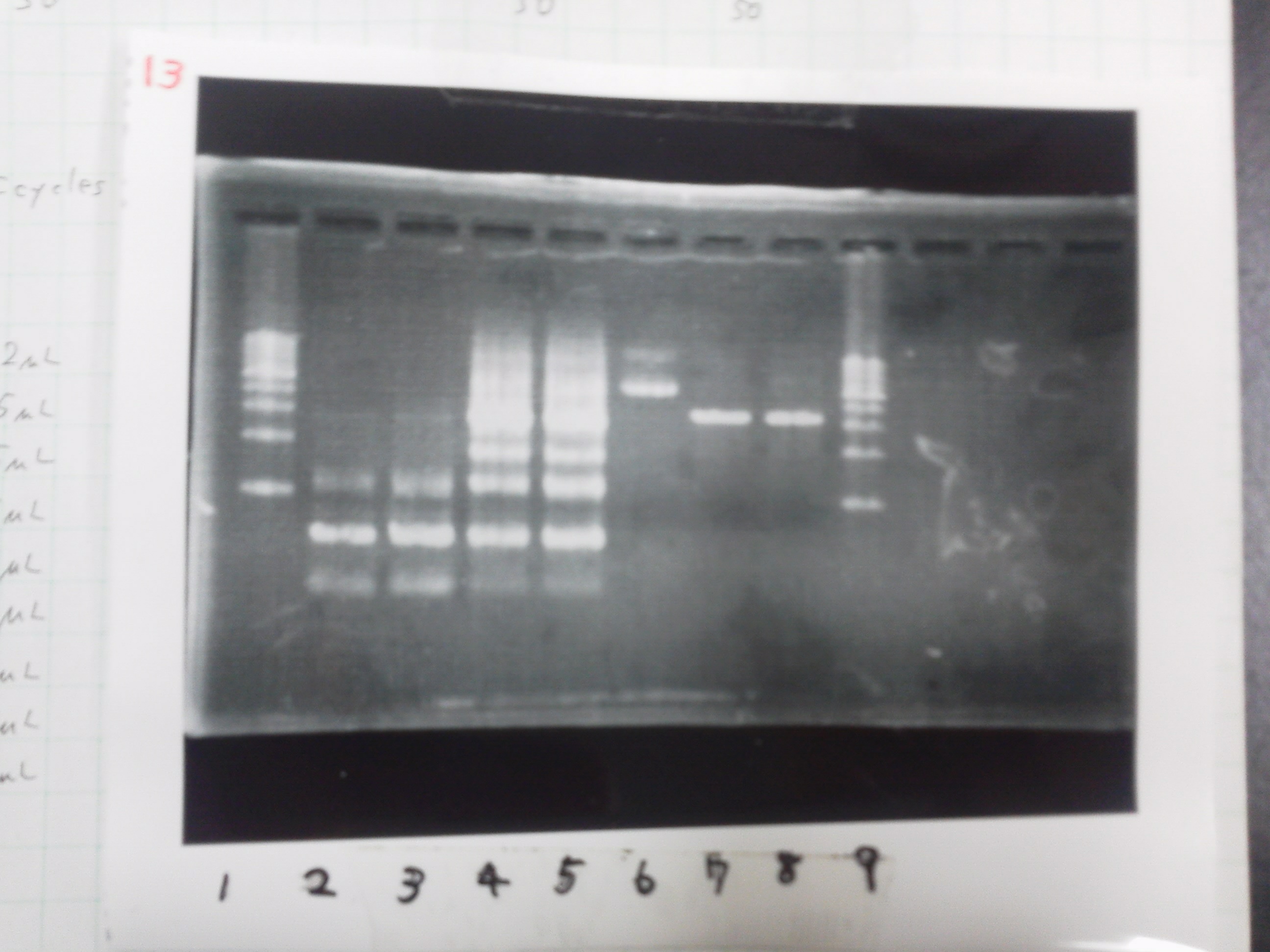
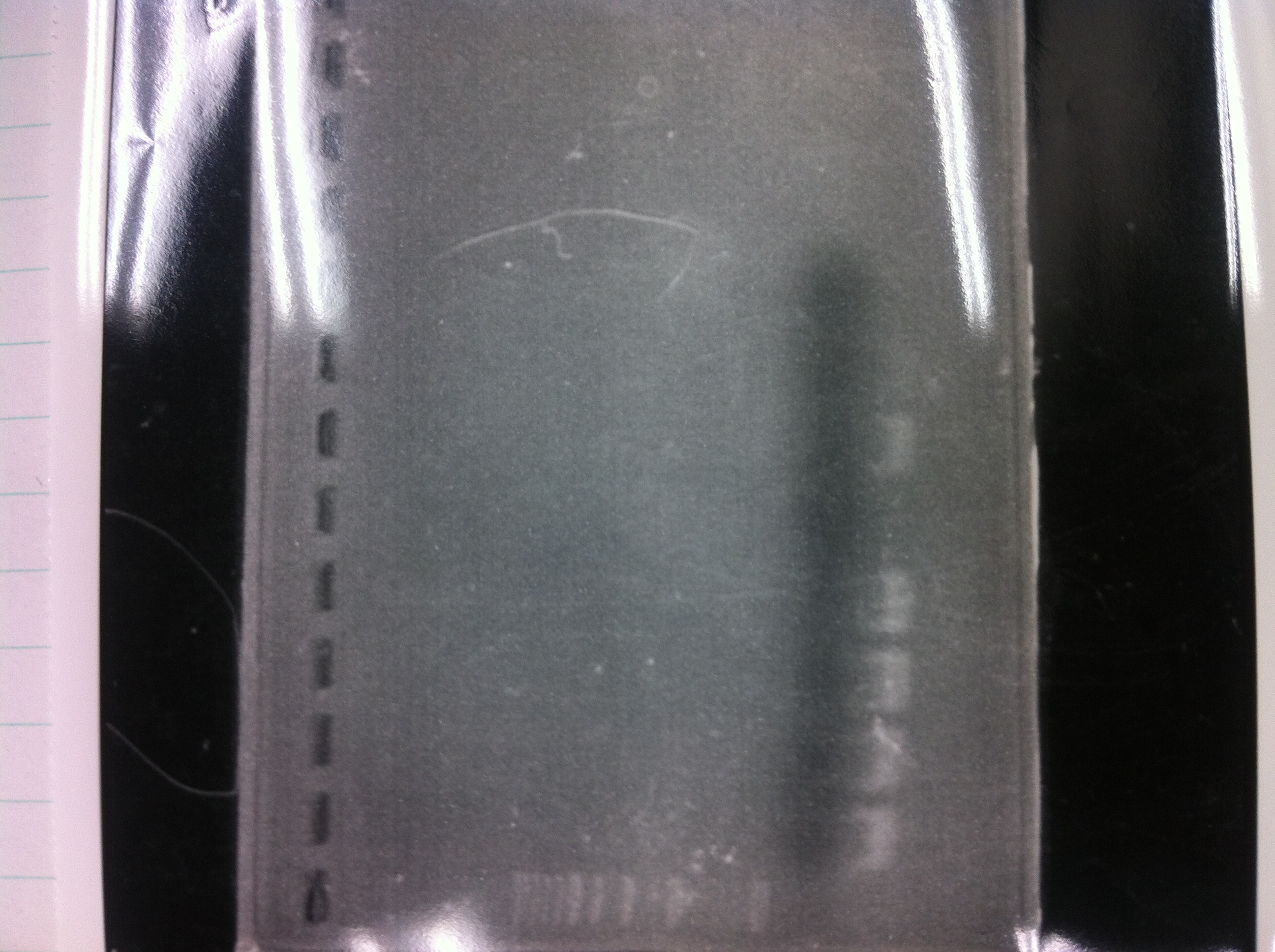
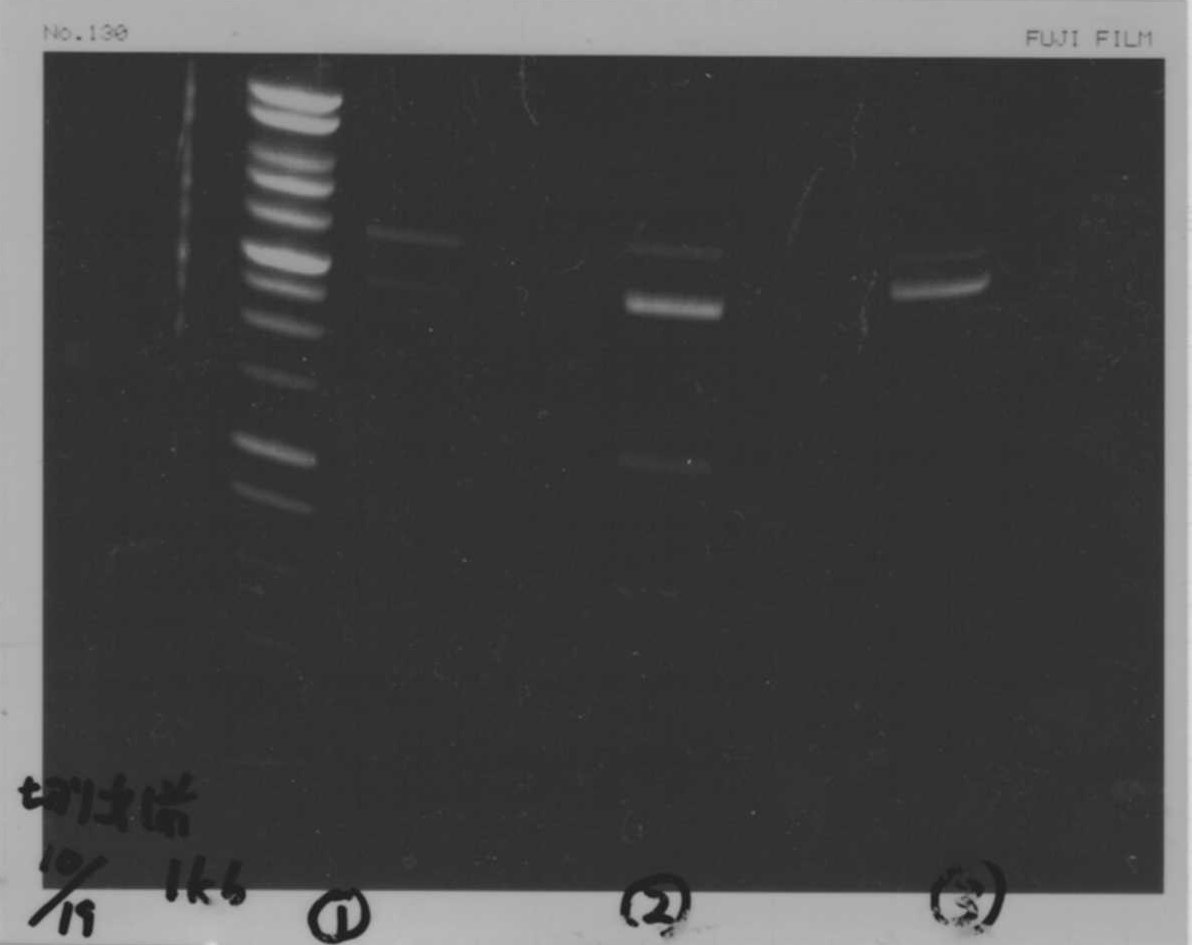
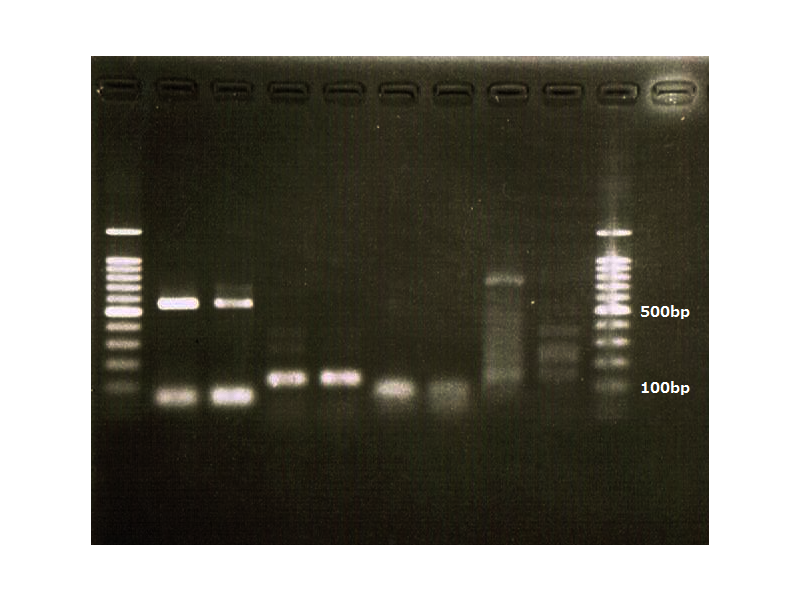
Lane1:Ladder 1kb
Lane2:T7-6His:FT about 2700bp
September 14
Ligation
Vector(pSB1C3: 12.5/µL, 2000bp) : 2µL = 19fmol
Insert(T7-R9-GFP-DT: 20.7ng/µL, 700bp ): 4µL = 180fmol
Ligation High ver.2 : 3µL
=> 16℃, 1 hours
Dpn1 processing
| PCR products(9/13) | Dpn1 | Total |
|---|---|---|
| 45 | 2 | 47 |
=>37℃, 1 hour
Self Ligation
| PCR products | Ligation High | T4 Kinase | MilliQ | Total |
|---|---|---|---|---|
| 2 | 5 | 1 | 7 | 15 |
=>16℃, 1 hour
Ligation
Vector(DT: 17.1/µL, 2100bp) : 2µL = 12fmol
Insert(T7-R9-FT: 20.1ng/µL, 650bp ): 5µL = 496fmol
Ligation High ver.2 : 3.5µL
=> 16℃, 1 hours
September 15
Restriction enzyme processing
| FT without RBS (34.6ng/µL) | EcoRI | PstI | 10× buffer H | MilliQ | total |
|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 2 | 7 | 20 |
Colony PCR
| Quick Tag | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
| 94℃ | 94℃ | 55℃ | 68℃ | cycles |
|---|---|---|---|---|
| 2min | 30sec | 30sec | 1min | 25 |
Electrophoresis
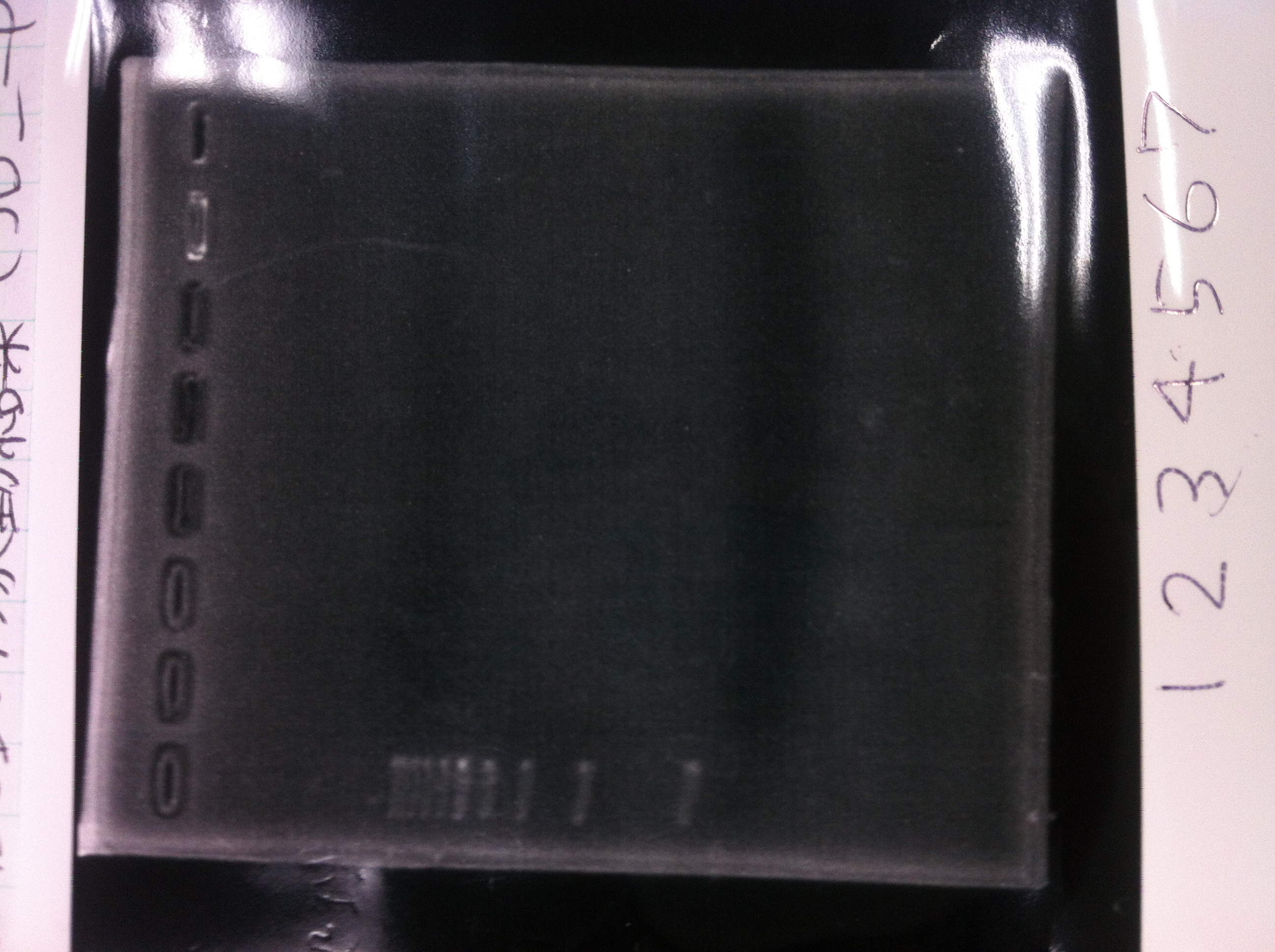
Lane1: ladder
Lane2: T7-R9-GFP-DT
Lane3: T7-R9-GFP-DT
Lane4: T7-R9-FT-DT
Lane5: T7-R9-FT-DT
Lane6: T7-R9-FT-DT
Lane7: T7-R9-FT-DT
Lane8: T7-R9-FT-DT
A member of secretion group electrophoresed DNA from Lane9 to Lane12.
Lane13: T7-His:FT
Lane14: T7-His:FT
Lane15: T7-His:FT
Lane16: T7-His:FT
Lane17: ladder
Liquid culture(3ml) 3:30~
T7-R9-FT-DT×2, T7-His:FT×2, T7-His:FT
September 16
Miniprep
①T7-R9-FT-DT 150ng/µL
②T7-R9-FT-DT 139ng/μL
③T7-R9-GFP-DT 67ng/µL
④T7-His:FT 153ng/μL
⑤T7-His:FT 73ng/μL
September 17
Purifying column
=>FT without RBS :36.4ng/µL
Ligation
Vector(PSB1C3: 12.5/µL, 2000bp) : 3µL = 9fmol
Insert(FT without RBS: 36.4ng/µL, 600bp ): 3µL = 92fmol
Ligation High ver.2 : 3µL
=> 16℃, 1 hours
September 18
Transformation by NAKAGAWA
| competent cell | DNA | plate |
|---|---|---|
| 20µL | PSB1C3 FT without RBS 1µL | CP+ |
Verification of R9 function by TAKEUCHI
| R9(20µg/µL) | 0.9µL |
| GFP(1.2mg/mL) | 2.23µL |
| RBS | 16.85µL |
| total | 20µL |
X5
Method:
1. Peel cuticles on parafilm by using the head of pencil.(Menasha,wI,54952)
2. Put plant cells into GFP&R9 or GFP for 5~30min.
3. Put plant cells into PBS.
4. Hoechst dyeing.
| 1 | 2 | 3 | 4 | 5 | 6 | |
| R9 | o | o | o | x | o | o |
| cuticle | o | o | o | o | x | x |
| soak in GFP | 5min | 15min | 30min | 5min | 5min | 30min |
September 19
Transformation(re) by NAKAGAWA,TAKEUCHI
competent cell:20µL
DNA(No RBS FT):1µL
plate :CP+
Transformation(re;re) by NAKAGAWA,TAKEUCHI
competent cell:10µL
DNA(No RBS FT):1µL
LB :100µL
plate :CP+
Adjusted GM Agar Medium making by TAKEUCHI
Ingredient of MS medium(SIGMA M5519): 0.88g
MES : 0.1g
ion exchanged water : 200mL
NaOH : 26µL(adjust to pH5.6)
Agar medium : 1.6g
Autoclave 120℃
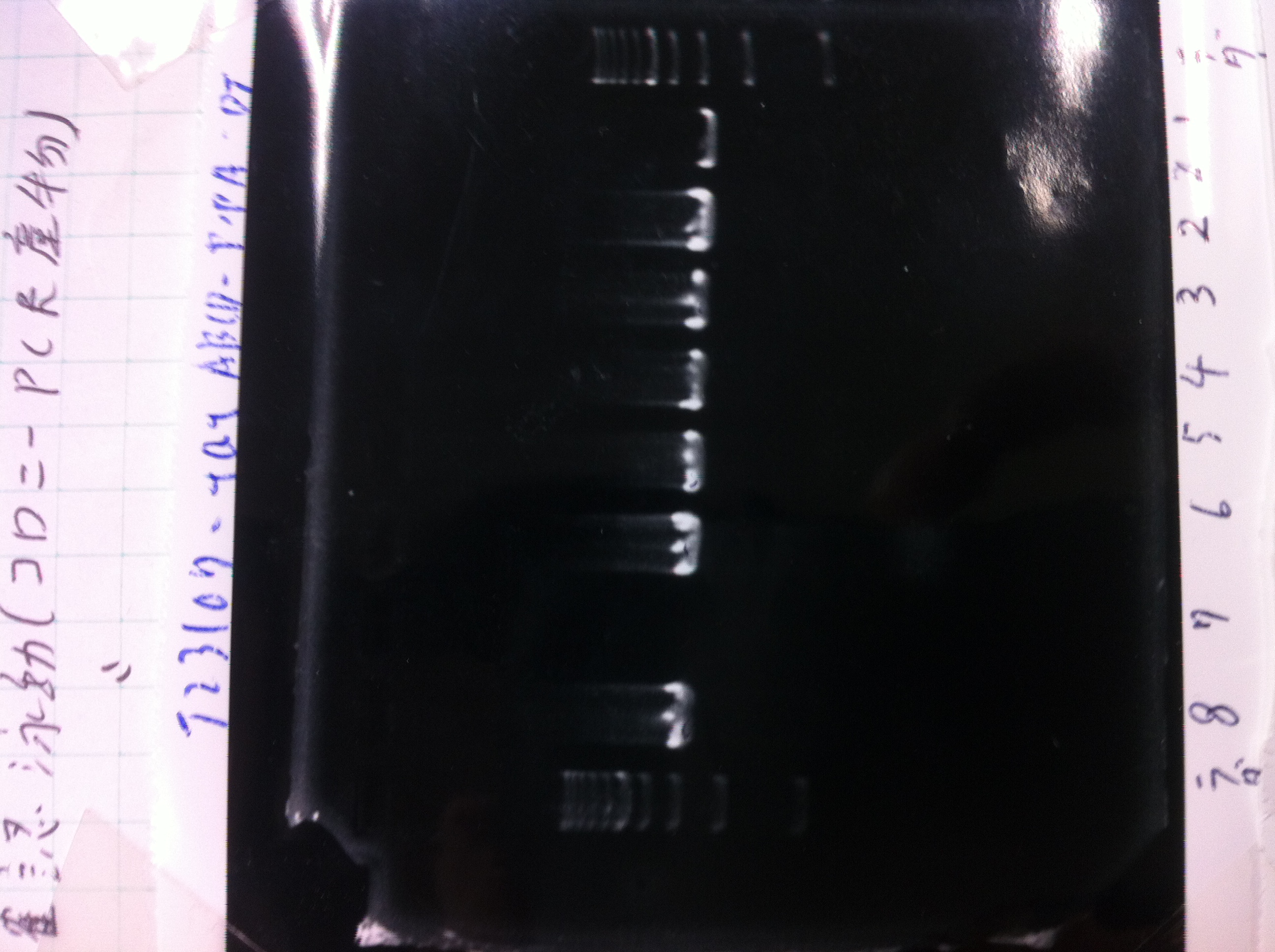
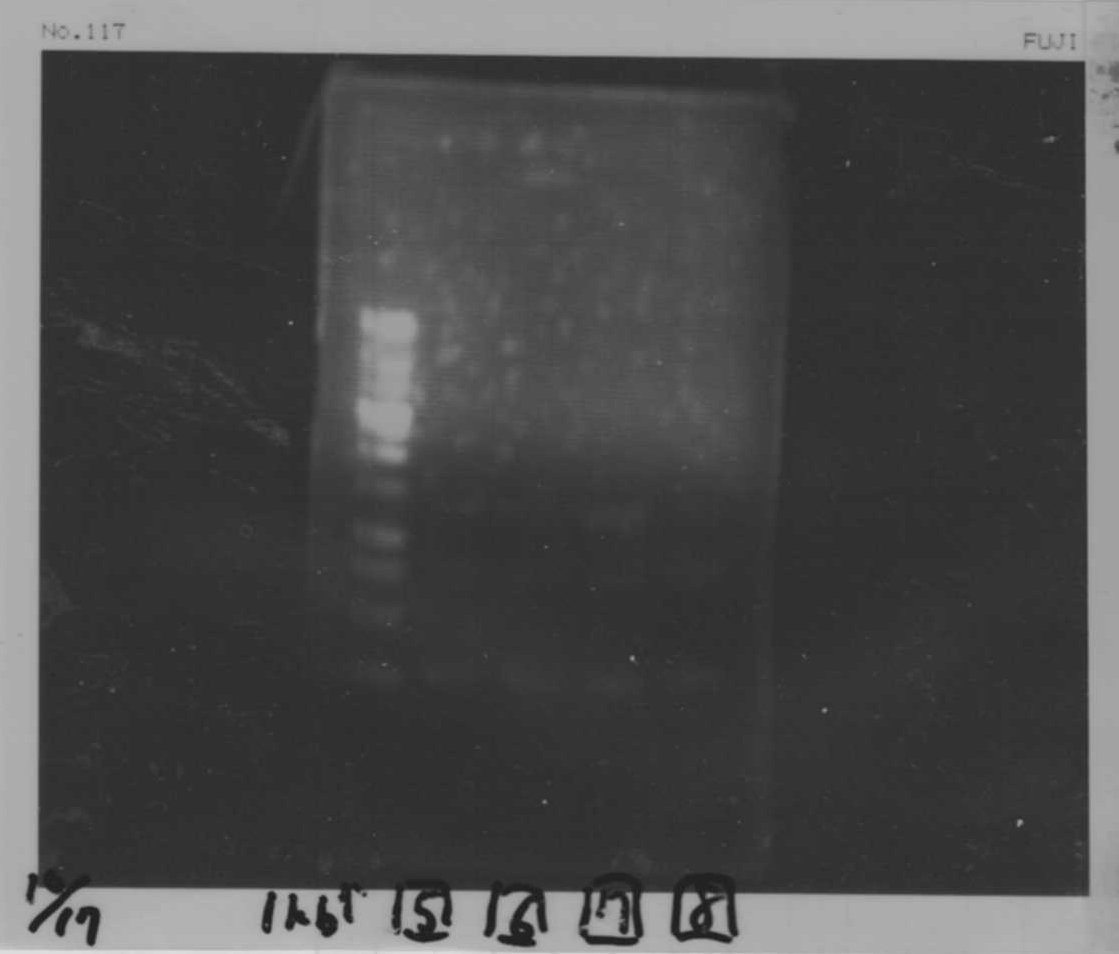
Colony PCR(re)
gelA
Lane1: 100bp Ladder
Lane2: No RBS FT colony number 9
Lane3: No RBS FT colony number 10
Lane4: No RBS FT colony number 11
Lane5: No RBS FT colony number 12
Lane6: No RBS FT colony number 13
Lane7: empty
Lane8: empty
gelB
Lane1: 100bp Ladder
Lane2: No RBS FT colony number 14
Lane3: No RBS FT colony number 15
Lane4: No RBS FT colony number 16
Lane5: No RBS FT colony number 17
Lane6: No RBS FT colony number 18
Lane7: No RBS FT colony number 19
Lane8: empty
gelC
Lane1: 100bp Ladder
Lane2: No RBS FT colony number 20
Lane3: No RBS FT colony number 21
Lane4: No RBS FT colony number 22
Lane5: No RBS FT colony number 23
Lane6: No RBS FT colony number 24
Lane7: empty
Lane8: empty
September 20
Liquid culture by NAKAGAWA
No RBS FT x8 (by using September 18)
Plate: CP+
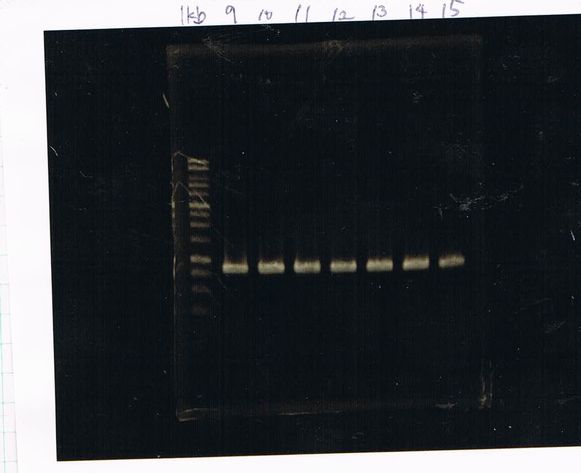
Colony PCR
No RBS FT by using September19
Lane1 : 100bp ladder
Lane2 : colony number 1
Lane3 : colony number 2
Lane4 : colony number 3
Lane5 : colony number 4
Lane6 : colony number 5
Lane7 : colony number 6
Lane8 : colony number 7
Lane9 : colony number 8
Lane10: empty
Lane11: empty
Lane12: 100bp ladder
Liquid culture
A-11, A-12
Miniprep
No RBS FT(by using September20)
1.-10.4µg/mL
2.-4.7µg/mL
3.-3.6µg/mL
4.-7.6µg/mL
5.-4.5µg/mL
6.-8.4µg/mL
7. 1.8µg/mL(average)
8. 18.1µg/mL
Restriction enzyme processing
| DNA(No RBS FT)x2 | E.coli | BufferH | MilliQ | total |
|---|---|---|---|---|
| 30µL | 1µL | 4µL | 5µL | 40µL |
in 37℃,2hours
Electrophoresis
DNA(No RBS FT) sample7,8: 10µL
Loading Dye : 2µL
Lane1: 1kb ladder
Lane2: empty
Lane3: No RBS FT(E) sample7
Lane4: empty
Lane5: No RBS FT(E) sample8
Lane6: empty
Electrophoresis(re)
RNA Extraction
5 leaves: 100mg
ISOGEN : 1mL
Elution : 20µL
cDNA Synthesis(1/4)
| gDNA wipeout buffer | template RNA | H2O | Total |
|---|---|---|---|
| 1µL | 0.5µL | 5.5µL | 7µL |
| Reverse Transcriptase | RT buffer | primer mix | template | Total |
|---|---|---|---|---|
| 0.5µL | 2µL | 0.5µL | 7µL | 10µL |
RT PCR
| buffer | dNTPs | MgSO4 | primer f | primer r | Template | KODplus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5 | 1.5 | 1 | 1 | 34.5 | 50 |
| 94℃ | 94℃ | 54℃ | 68℃ | cycles |
|---|---|---|---|---|
| 2min | 15sec | 30sec | 10sec | 30 |
1.TUBULIN
2.FUL
3.SEP3
4.AP1
September 22
mRNA extraction
FT and R9
Arabidopsis thaliana's leaves : 200mg
| R9 | FT | total |
|---|---|---|
| 1.575μL | 33.425μL | 35μL |
| R9 | GFP | |
|---|---|---|
| 1.575 | 33.425 | 35 |
1. Strip cuticles on palafilm by the head of a pencil.
2. Soak plant cells in R9 & FT or R9 & GFP for 2 hours(19:45~).
3. Add PBS (21:45~)
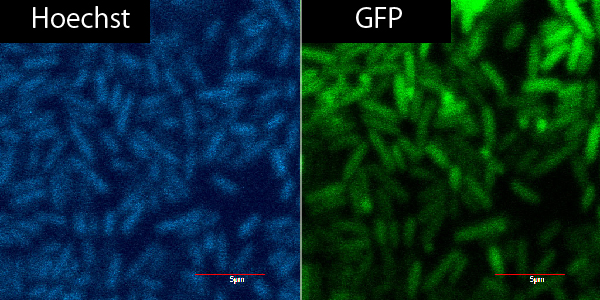
Microscope
R9+GFP+Plant Cell ver.3
R9+
| R9 | GFP | PBS | total |
|---|---|---|---|
| 0.9 | 2.25 | 16.85 | 20μL |
R9-
| R9 | GFP | PBS | total |
|---|---|---|---|
| 0 | 2.25 | 17.65 | 20 |
same as 9/20, only not on parafilm but on microscope slide from the start.
September 23
RNA Extraction
FT- (GFP+)
FT+
| ISOGEN | chloroform | elution | total |
|---|---|---|---|
| 1 | 200 | 11 | 212 |
Reverce Transcription
| gDNA wipeout buffer | RNA | total |
| 1 | 6 | 7 |
at 42 degree, for 2min.
| Reverce Transcriptase | RT buffer | primer mix | Template | total |
|---|---|---|---|---|
| 0.5 | 2 | 0.5 | 7 | 10 |
at 42 degree, for 30min.
at 95 degree, for 3min.
RT-PCR
| Buffer | dNTPs | MgSO4 | primer | Template | KOD Plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5 | 1 | 1 | 34.5 |
94℃, 2min
94℃, 15sec
54℃, 30sec
68℃, 10sec
1.NC TUBULIN
2. FUL
3. SEP3
4. AP1
5.FT TUBULIN
6. FUL
7. SEP3
8. AP1
September 24
ScreeningPCR & Electrophoresis
Lane
1. 100bp ladder
2. T7-His:FT ① 9.2ng/μL
3. T7-His:FT ② 11.7ng/μL
4. T7-His:FT ③ 23.6ng/μL
5. T7-His:FT ④ 21.6ng/μL
6. T7-His:FT ⑤ 30.6ng/μL
7. T7-His:FT ⑥ 51.5ng/μL
8. T7-His:FT ⑦ 47.1ng/μL
9. T7-His:FT(A4) 20.0ng/μL
10. T7-His:FT(B1) 18.8ng/μL
11. final2(by_Secretion_group)
12. blank
FT Introduce
| FT | R9 | PBS | total |
|---|---|---|---|
| 50 | 2.88 | 11.12 | 64 |
| GFP | R9 | PBS | total |
|---|---|---|---|
| 7.2 | 2.88 | 53.02 | 64 |
We took 1 leaf out of 6 individuals of Arabidopsis thaliana that grow three weeks one by one
Leaves 30mg
1.cut off leaves
2.Inject the juice(by terumo-syringe)(Center tip、tuberculin 1nl)
3.store 20 min, then add PBS 600μL
September 25
RNA extraction
FT- (GFP+) FT+
elution 6µL
Reverce Transcription
| gDNA wipeout buffer | RNA | total |
| 1 | 6 | 7 |
at 42 degree, for 2min.
| Reverce Transcriptase | RT buffer | primer mix | Template | total |
|---|---|---|---|---|
| 0.5 | 2 | 0.5 | 7 | 10 |
at 42 degree, for 30min.
at 95 degree, for 3min.
RT-PCR
| Buffer | dNTPs | MgSO4 | primer | Template | KOD Plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5 | 1 | 1 | 34.5 |
94℃, 2min
94℃, 15sec
54℃, 30sec
68℃, 10sec
1.TUBULIN NC
2.TUBULIN FT
3.FUL NC
4.FUL FT
5.SEP3 NC
6.SEP3 FT
7.AP1 NC
8.AP1 FT
October 12
Transformation
| BL21(DE3) | T7-His-FT | T7-His-GFP-DT | total |
|---|---|---|---|
| 1μL | 10μL | 11μL | |
| 1μL | 10μL | 11μL |
October 16
Liquid culture
T7-His-FT 3mL
Transformation (retry)
| BL21(DE3) | T7-His-GFP-DT | total |
|---|---|---|
| 1μL | 10μL | 11μL |
October 17
Moved 1mL of Liquid culture of T7-His-FT in 100mL of SOC medium -> 2 samples
Added IPTG 100μL to the medium(final conc. is 1mM) when value of OD600 is 0.649
Incubated at 20℃ for 4 hours.
Collect bacterial cells and freeze-preserved.
October 20
Liquid culture
2mL (T7-His-GFP)
October 21
Collect bacterial cells same way described above(value of OD600 is 0.8, at 37℃, for 4 hour)
Purification of GFP
| Lysis buffer | lysozyme | Triton X-100 | Ni-NTA resin | Elution buffer |
|---|---|---|---|---|
| 5mL | 0.1mL | 0.3mL | 1mL | 2.5mL |
fraction1:31.5µM->After consentrating 47.4µM
fraction2:15.8µM->After consentrating 33.4µM
fraction3:20.1µM
(Extinction coefficient=21,890)
RNA Extraction
Added 3μL of IPTG to 3mL of E.coli and then incubated 2 hour.
| ISOGEN | chloroform | isopropanol | Elution buffer |
|---|---|---|---|
| 1mL | 0.2mL | 0.5mL | 11μL |
deluted to 250ng/μL
Reverce Transcription
| RNA(250ng/μL) | gDNA wipe out buffer | dH2O | total |
|---|---|---|---|
| 0.5μL | 1μL | 5.5μL | 7μL |
| Template | RT buffer | Primer mix | RT | total |
|---|---|---|---|---|
| 7μL | 2μL | 0.5μL | 0.5μL | 10μL |
RT-PCR
Template is 20 times deluted cDNA
| buffer | dNTPs | MgSO4 | primer | primer | Template | KOD-Plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5(His-FT f) | 1.5(FT sequence r) | 1 | 1 | 33 | 50 |
| 5 | 5 | 2 | 1.5(GAPDH) | 1 | 1 | 34.5 | 50 |
94℃ 2min
94℃ 15sec
55℃ 30sec
68℃ 30sec
for 30 cycles
lane_1 : GAPDH
lane_2 : FT
lane_3 : 100bp ladder
October 22
Restriction
| T7-His-FT(A4, 20ng/μL) | T7-His-FT(B2, 73ng/μL) | bufferH | EcoR1 | Pst1 | MilliQ | total |
|---|---|---|---|---|---|---|
| 10μL | 2μL | 0.5μL | 0.5μL | 7μL | 20μL | |
| 5μL | 2μL | 0.5μL | 0.5μL | 12μL | 20μL |
at 37℃ for 1 hour
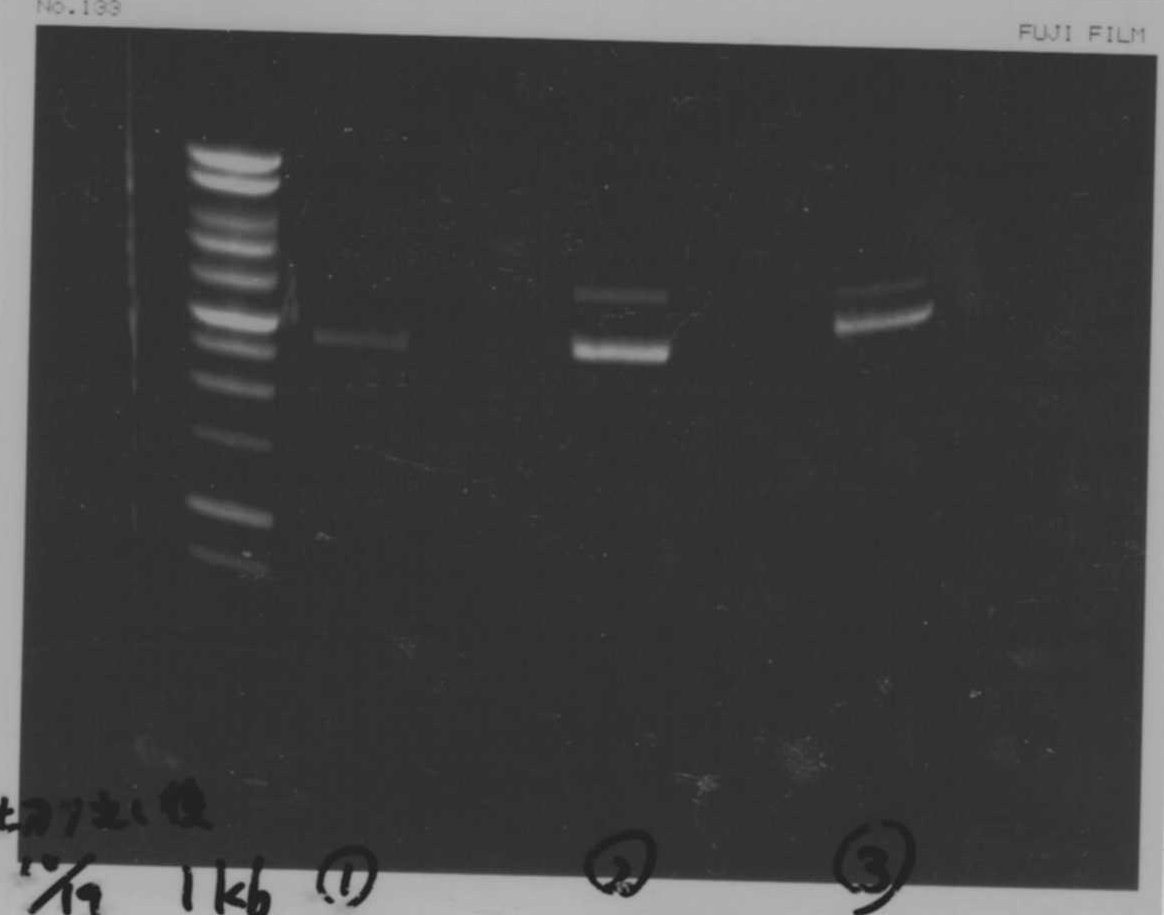
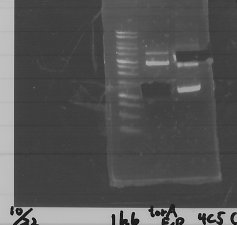
lane1 : T7-His-FT(B2)
lane2 : T7-His-FT(B2, EcoR1/Pst1)
lane3 : T7-His-FT(A4)
lane4 : T7-His-FT(A4, EcoR1/Pst1)
Transformation
| T7-His-FT(B2) | BL21(DE3) | total |
|---|---|---|
| 1 | 10 | 11 |
October 23
RNA Extraction from plant cell
| ISOGEN | chloroform | isopropanol | High salt solution | Elution buffer |
|---|---|---|---|---|
| 1mL | 0.25mL | 0.25mL | 0.25mL | 11μL |
notes: After the homogenization with ISOGEN, sample 1 was incubated at 50℃ for 10min.
Sample1. 855.8ng/µL
Sample2. 812.4ng/µLL
October 24
Collect bacterial cells(T7-6His:FT) same way described above(value of OD600 is 0.722, at 37℃, for 4 hour)
2mL of culture was used for RNA extraction.
Purification of FT
| Lysis buffer | lysozyme | Triton X-100 | Ni-NTA resin | Elution buffer |
|---|---|---|---|---|
| 5mL | 0.15mL | 0.3mL | 0.5mL | 1.2mL |
fraction1:65.1µM
fraction2:42.1µM
fraction3:10.9µM
fraction4:6.8µM
(Ectinction coefficient=21,430)
RNA extraction
| ISOGEN | chloroform | isopropanol | Elution buffer |
|---|---|---|---|
| 1mL | 0.2mL | 0.5mL | 11μL |
diluted to 250ng/μL
Reverce Transcription
| RNA(250ng/μL) | gDNA wipe out buffer | dH2O | total |
|---|---|---|---|
| 0.5μL | 1μL | 5.5μL | 7μL |
| Template | RT buffer | Primer mix | RT | total |
|---|---|---|---|---|
| 7μL | 2μL | 0.5μL | 0.5μL | 10μL |
RT-PCR
Template is 20 times deluted cDNA
| buffer | dNTPs | MgSO4 | primer | primer | Template | KOD-Plus | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5(His-FT f) | 1.5(FT sequence r) | 1 | 1 | 33 | 50 |
| 5 | 5 | 2 | 1.5(GAPDH) | 1 | 1 | 34.5 | 50 |
94℃ 2min
94℃ 15sec
55℃ 30sec
68℃ 30sec
for 30 cycles
lane_1 : 100bp ladder
lane_2 : GAPDH
lane_3 : FT
Assay of FT activation
| protein-R9 solution mix | FT | GFP | R9 | PBS |
|---|---|---|---|---|
| sample1 (FT-) | - | 200µL | 0.11mg | 100µL |
| sample2 (FT+) | 230µL | - | 0.21mg | 70µL |
100mg of Arabidopsis thaliana leaves were harvested for each samples and injected the solution mix.
incubation at room temperature for 13h in dark
October 25
| ISOGEN | chloroform | isopropanol | High salt solution | Elution buffer |
|---|---|---|---|---|
| 1mL | 0.25mL | 0.25mL | 0.25mL | 11μL |
Sample1. 682.7ng/µL
Sample2. 859.3ng/µLL
diluted to 250ng/μL
Reverce Transcription
| RNA(250ng/μL) | gDNA wipe out buffer | dH2O | total |
|---|---|---|---|
| 1μL | 2μL | 11μL | 14μL |
| Template | RT buffer | Primer mix | RT | total |
|---|---|---|---|---|
| 14μL | 4μL | 1μL | 1μL | 20μL |
RT-PCR
Template is 20 times diluted cDNA
| buffer | dNTPs | MgSO4 | primer | Template | KOD-Plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 2 | 1.5 | 1 | 1 | 34.5 | 50 |
94℃ 2min
94℃ 15sec
54℃ 30sec
68℃ 10sec
February 7
Preculture
We started preculture at 12:10.
February 8
Liquid culture
We start culturing with 300mL of LB medium.
| time | OD600 |
|---|---|
| 12:00 | start |
| 14:10 | 0.019 |
| 14:45 | 0.154 |
| 15:05 | 0.267 |
| 15:21 | 0.64 |
Making Competent Cell
We made competent cells.
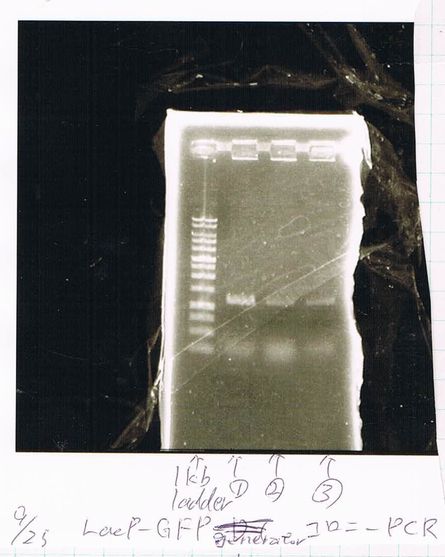
Transformation
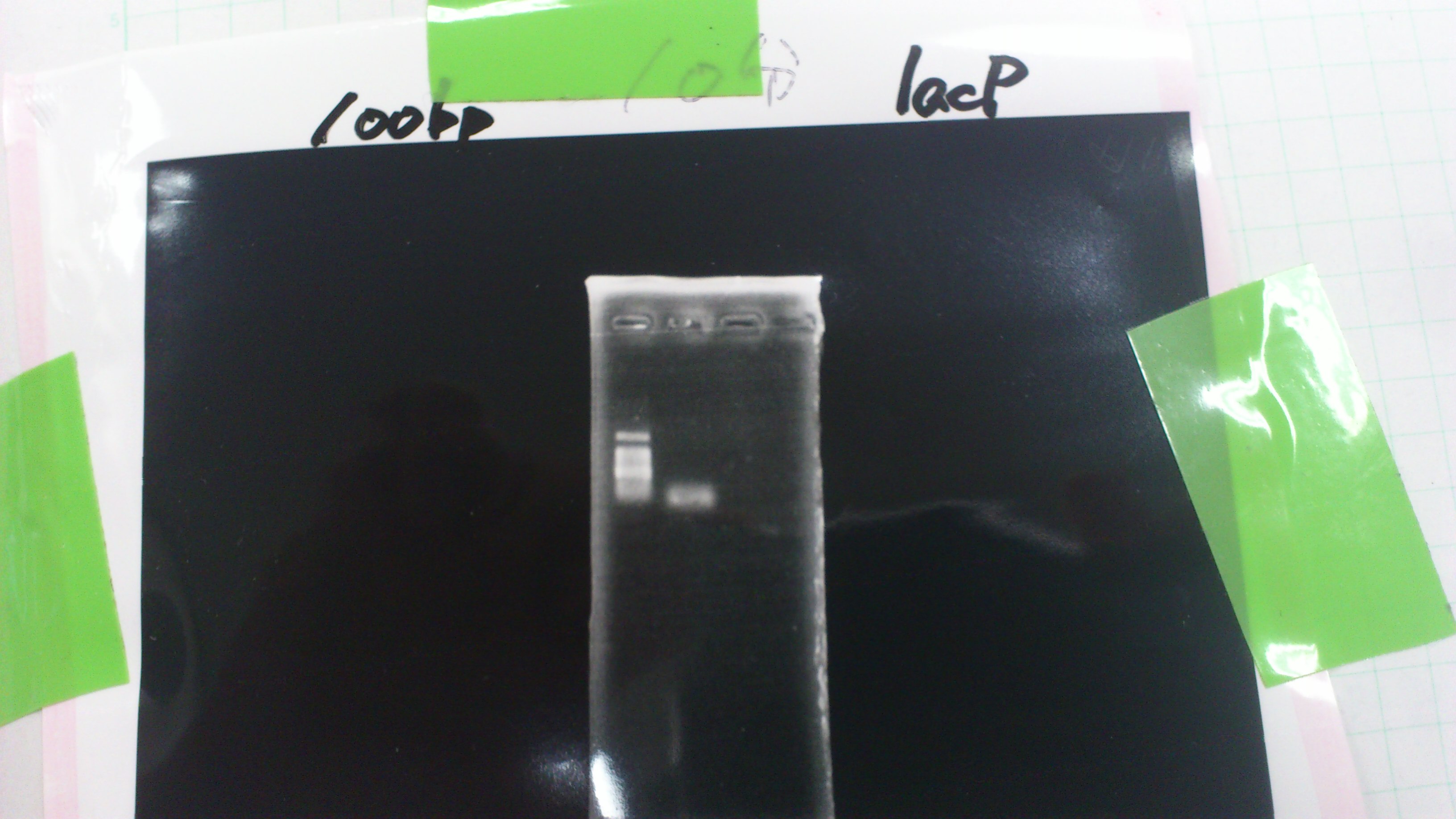
pGEM_TAP
LacP (BBa_R0011)
DT (BBa_B0015)
Making Culture Medium Plates
We made 200mL of ampicillin culture, kanamycin culture, and chloramphenicol culture.
Transformation
GFP(BBa_E0040) in pSB1A2
DT(BBa_B0015) in pSB1AK3
ara(BBa_I0500) in pSB2K3
LacP(BBa_R0011) in pSB1A2
February 9
Transformation
BBa-E0040(GFP)(Mr.Fujita)
Liquid culture
DT, LacP colony transformed on February 8
colony of competent cell made on February 8
B0040 1.4k pSB1A2 B0034 1.2M pSB1A2(from iGEM parts plate)
Making Competent cells
We did preculture for overnight. We put 1.5mL of preculture on 150mL of LB culture.
| time | OD600 |
|---|---|
| 11:45 | start |
| 13:30 | 0.048 |
| 14:30 | 0.168 |
| 15:03 | 0.256 |
| 15:20 | 0.405 |
| 15:35 | 0.459 |
| at last | 0.576 |
February 11
Checking Transformation efficiency
Conpetent cell's transformation efficiency is 1.3x10^4colonys/μg
February 13
Transformation
Const promoter J23110, J23109, J23100
| DNA | Competent cell | Total |
|---|---|---|
| 1μL | 20 | 21 |
No colony was there on February 14
Liquid culture
LacP, DT, RBS(BBa_B0034),GFP
start at 20:00
in Plus grow with Ampicilin 3mL
February 14
Miniprep
| DNA | concentration[μg/μL] |
| LacP3 | 39.6 |
| LacP4 | 40.8 |
| LacP5 | 28.9 |
| RBS1 | 28.2 |
| RBS2 | 57.4 |
| RBS3 | 13.2 |
| DT3 | 69.7 |
| DT4 | 64.4 |
| DT5 | 61.5 |
| GFP1 | 64.0 |
| GFP2 | 50.5 |
| GFP3 | 66.0 |
Restrictive Digestion
Const promoter J23100
| DNA | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for overnight
February 15
Making gel
1% Agarose gel
| Agarose | TAE |
|---|---|
| 1.6g | 160mL |
Electrophoresis
Gel No.1
| Restriction product | loading dye |
|---|---|
| 5μL | 1 |
The marker was 1kb ladder
It seemed that this restriction product was not cut.
Gel No.2
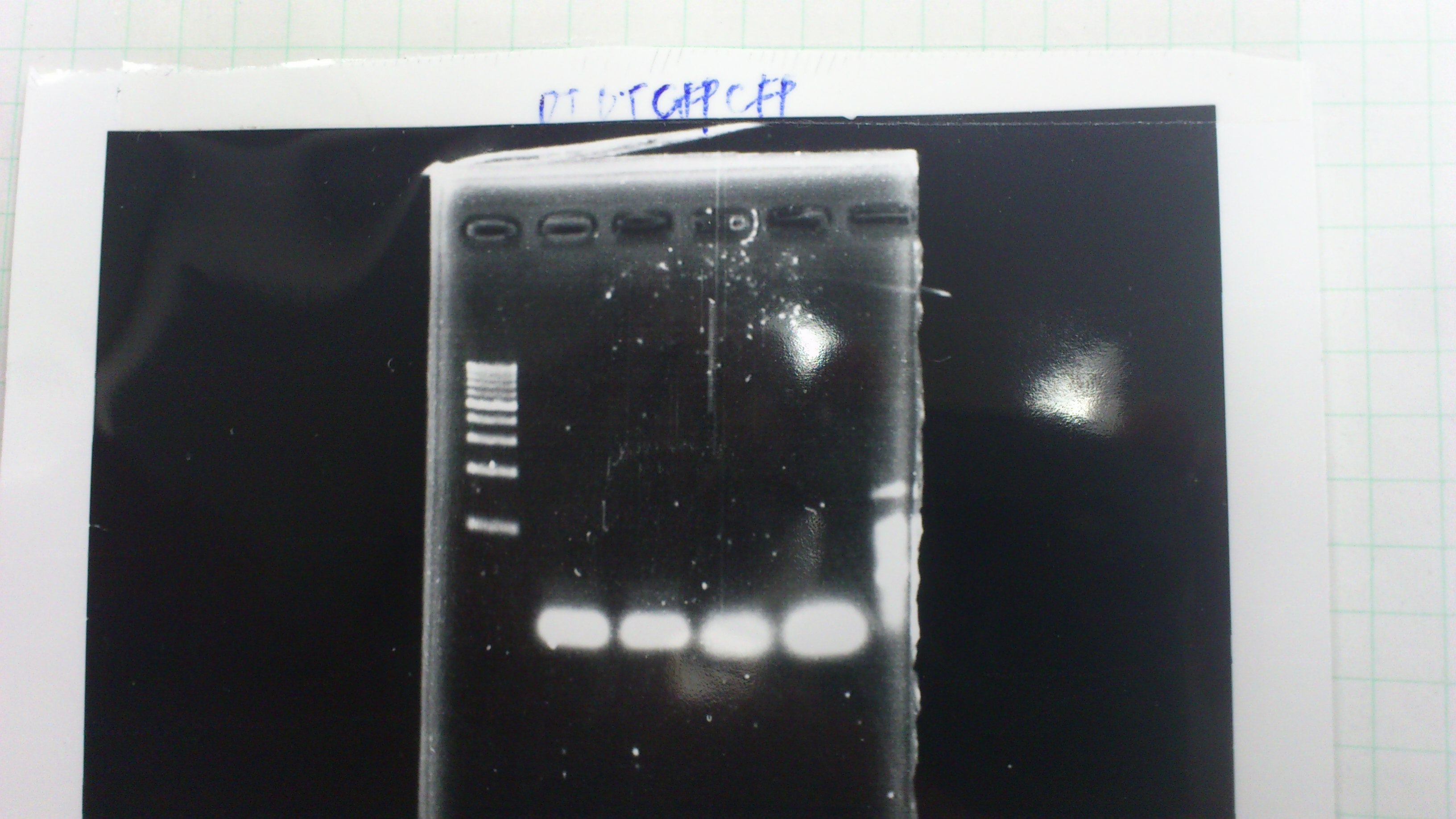
Lane1 : 1kb ladder
Lane2 : J23100 2μL + 6*Loading dye 1μL
Lane3 : J23100(Spe1,Pst1) 5μL
Lane4 : J23100(Spe1,Pst1) 2μL
- There were bands on lane_2 and we cannot identify these bands because the sample of lane_2 was not cut with any restriction enzyme.
- There must have been bands at 2100bp and 883bp on lane_3 and lane_4.
Testing whether restriction enzyme were deactivated or not
| DNA(DT) | restriction enzyme | Buffer | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 0.5 | 3 | 0.5 | 16 | 30 |
at 37℃ for Oveernight
Restriction enzyme means Spe1(1, 2) Pst1(1, 2, 3) in this time.
February 16
Electrophoresis
1. 1kb ladder
2. DT2
3. DT3
4. DT2 (Spe1-1)
5. DT2 (Spe1-2)
6. DT2 (Pst1-1)
7. DT2 (Pst1-2)
8. DT2 (Pst1-3)
9. DT3 (Pst1-4)
10. 1kb ladder
Pst1-1, Pst1-2, and Pst1-3 did not cut DNAs. They seemed to be deactivated.
Genomic PCR
| 10*Buffer for KOD Plus | 2mM dNTPs | 25mM MgSO4 | 10μM primer-f | 10μM primer-r | 158ng/μL Genomic DNA | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
Electrophoresis
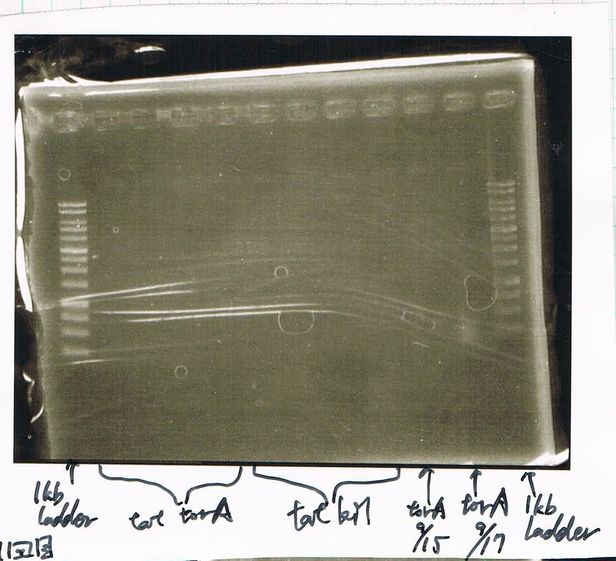
1. 1kb ladder
2. TatABCD (2.5kb)
3. TAMO reductase (2.7kb)
4. Negative control
We got bands of TatABCD but there were nonspecific amplification products.
We failed amplification of TAMO reductase.
Transformation
Constitutive promoter (BBa_J23107, BBA_J23117)
High copy plasmid (pSB1AT3)
| DNA | competent cell |
|---|---|
| 1μL | 10μL |
February 17
PCR
We did PCR to amplify products of PCR that we had done yesterday but we could not amplify TatABCD.
Genomic PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 50 |
Predenature 94℃, 2min
Denature 98℃, 10sec
Annealing 57℃, 30sec
Extension 68℃, 2.5min
(30cycles)
Electrophoresis
1. 1kb ladder
2. TAMO reductase
3. Negative control
Restrictive Digestion
| J23100 | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
Genomic PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|
| TMAO reductase | 2.5 | 2.5 | 3 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| TatABCD | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| TatABCD | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 5 | 0.5 | 10.5 | 25 |
Electrophoresis
1. 1kb ladder
2.3. TAMO reductase
4. TatABCD
5. TatABCD(10 times amount of genome)
Checking of restriction enzyme
| DT | enzyme | Buffer | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 2 | 0.5 | 3 | 0.5 | 24 | 30 |
at 37℃ for overnight
We checked EcoR1 and Xba1.
February 18
Miniprep
| μg/mL | 260/280 | 230/260 | |
|---|---|---|---|
| JS3117-1 | 135 | 1.5 | 2.06 |
| JS3117-2 | 75 | 1.6 | 1.63 |
| JS3109-1 | 115 | 1.5 | 1.88 |
| JS3109-2 | 75 | 1.65 | 1.71 |
| pSB1AT3-1 | 70 | 1.66 | 1.83 |
| pSB1AT3-2 | 100 | 1.52 | 1.54 |
diluted to 25 times
Making Competent cells
We put 3mL of preculture product on yesterday onto 300mL of LB medium
| time | OD600 |
|---|---|
| 10:30 | start |
| 12:10 | 0.118 |
| 13:00 | 0.270 |
| 13:30 | 0.502 |
Transformation
| pSB1AT3-2 | competent cell | MilliQ | Total |
|---|---|---|---|
| 0 | 20 | 10 | 30 |
| 2 | 20 | 8 | 30 |
| 10 | 20 | 0 | 30 |
- Results(on Feb. 19)
| pSB1AT3-2 | number of colony |
|---|---|
| 0 | 0 |
| 2 | 177 |
| 10 | 590 |
Transformation efficiency 7.4x10^4 colonys/μg
February 20
Restrictive Digestion
sample 1
| DT plasmid | EcoR1 | Xba1 | Buffer | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 7.5 | 0.5 | 0.5 | 3 | 0.5 | 18 | 30 |
sample2
| GFP plasmid | EcoR1 | Spe1 | Buffer | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
Electrophoresis
- sample1
a. sample1 2μL + MilliQ 3μL + 6×Loading Dye 1μL
b. sample1 5μL + 6×Loading Dye 1μL
lane1. 1kb ladder
lane2. a
lane3. b
lane4. a
lane5. b
lane6. a
lane7. b
lane8. 1kb ladder
- sample2
c. sample2 2μL + MilliQ 3μL + 6×Loading Dye 1μL
d. sample2 5μL + 6×Loading Dye 1μL
lane1. 1kb ladder
lane2. c
lane3. d
lane4. 1kb ladder
PCR
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | genomic DNA | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|
| TatABCD1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
| TatABCD2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
| TMAO reductase1 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0.5 | 0.5 | 15.5 | 25 |
| TMAO reductase2 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 5 | 0.5 | 10.5 | 25 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 2.5min
→30cycles
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | PCR products | genomic DNA | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| TatABCD1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0 | 0.5 | 0.5 | 16 | 25 |
| TatABCD2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 1 | 0 | 0.5 | 15.5 | 25 |
| TMAO reductase1 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0 | 0 | 0.5 | 16 | 25 |
| TMAO reductase2 | 2.5 | 2.5 | 2 | 0.75 | 0.75 | 0 | 0 | 0.5 | 16 | 25 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 2.5min
→35cycles
Electrophoresis
lane1.1kb ladder
lane2.TatABCD
lane3.TatABCD
lane4.TAMO1
lane5.TAMO2
lane6.1kb ladder
lane1.1kb ladder
lane2.TatABCD1
lane3.TatABCD2
lane4.TAMO
lane5.TAMO
lane6.1kb ladder
February 21
PCR (Advantage HF protocol)
| buffer | dNTPs | primer-f | primer-r | gDNA | PCR products | DW | polymerase | Total | |
|---|---|---|---|---|---|---|---|---|---|
| TatABCD | 2.5 | 2.5 | 0.75 | 0.75 | 1 | 0 | 17 | 0.5 | 25 |
| TMAO | 2.5 | 2.5 | 0.75 | 0.75 | 0 | 1 | 17 | 0.5 | 25 |
Predenature 94℃ 1min
Denature 94℃ 30sec
Annealing 58℃ 30sec
Extension 68℃ 3min
→25cycles
Electrophoresis
1. 1kb ladder 2μL
2. TatABCD 5μL + 6×Loading Buffer 1μL
3. TAMO 5μL + 6×Loading Buffer 1μL
4. 1kb ladder 2μL
Liquid culture
pSB3C5-1,2
pSB4K5-1,2
February 22
Gel extraction
lane 1 of the gel 45.0μg/mL
PCR purification
product 38.2μg/mL
PCR
TMAO reductase
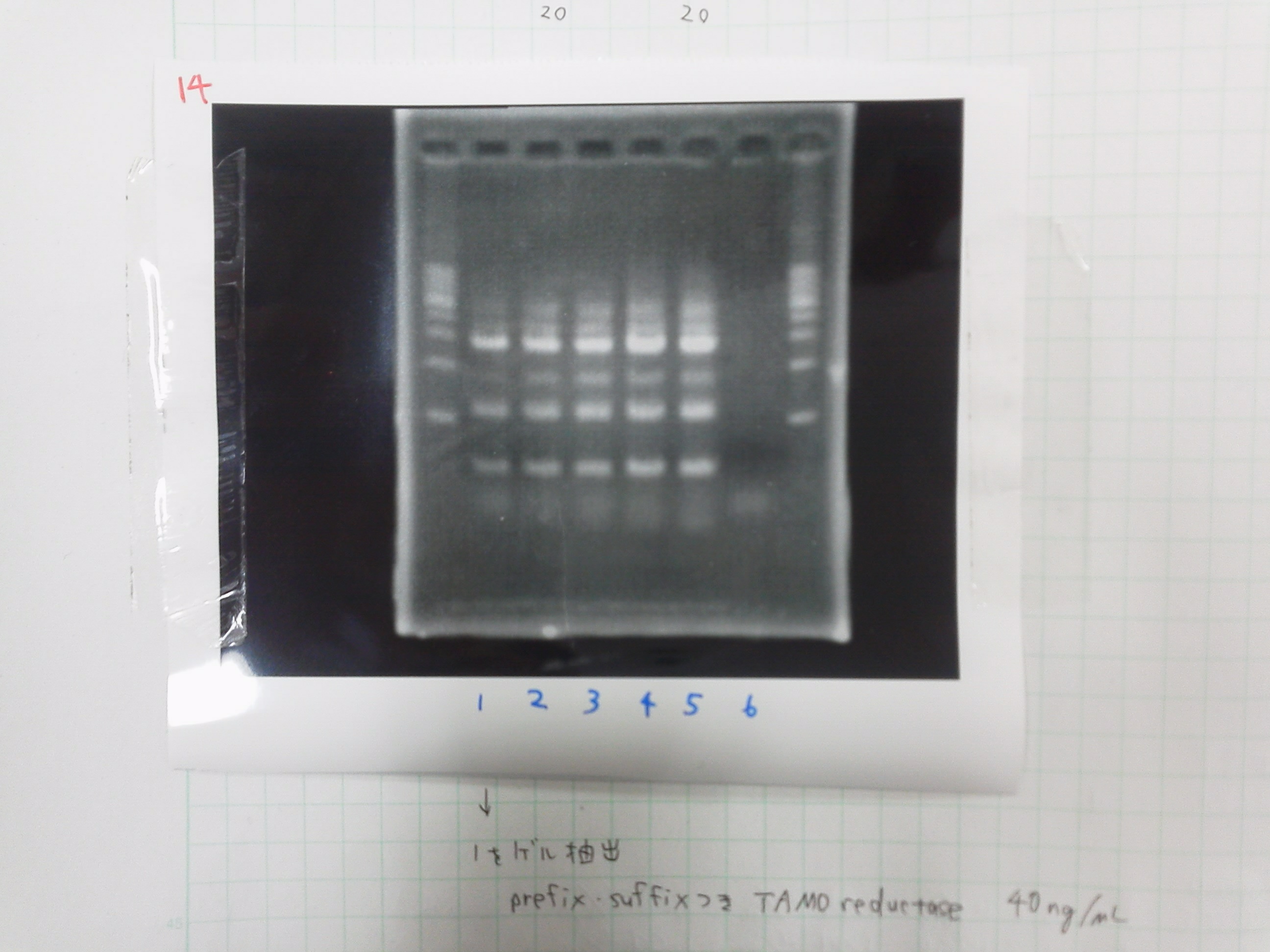
| Buffer | dNTPs | MgSO4 | prefix primer-f | suffix primer-r | product of gel extract | product of PCR purification(1ng/μL) | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 4 | 1.5 | 1.5 | 0.5 | 0 | 1 | 32.5 | 50 |
| 2 | 5 | 5 | 4 | 1.5 | 1.5 | 1 | 0 | 1 | 32.5 | 50 |
| 3 | 5 | 5 | 4 | 1.5 | 1.5 | 0 | 0.5 | 1 | 32.5 | 50 |
| 4 | 5 | 5 | 4 | 1.5 | 1.5 | 0 | 1 | 1 | 32.5 | 50 |
94℃, 2min
98℃, 10sec
59℃, 30sec
68℃, 3min
→25cycles
Electrophoresis
1. 1kb ladder
2. TAMO1
3. TAMO2
4. TAMO3
5. TAMO4
6. constructive promoter 1-18C
7. constructive promoter Spe1
8. constructive promoter Pst1
9. 1kb ladder
PCR
| Buffer | dNTPs | MgSO4 | prefix primer-f | suffix primer-r | product of PCR purification(1ng/μL) | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 1 | 32.5 | 50 |
| 2 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
| 3 | 5 | 5 | 3 | 1.5 | 1.5 | 2 | 1 | 31 | 50 |
| 4 | 5 | 5 | 3 | 1.5 | 1.5 | 3 | 1 | 30 | 50 |
| 5 | 5 | 5 | 3 | 1.5 | 1.5 | 10 | 1 | 29 | 50 |
| 6 | 5 | 5 | 3 | 1.5 | 1.5 | 0 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
59℃, 30sec
68℃, 3min
→25cycles
Electrophoresis
Checking Dpn1
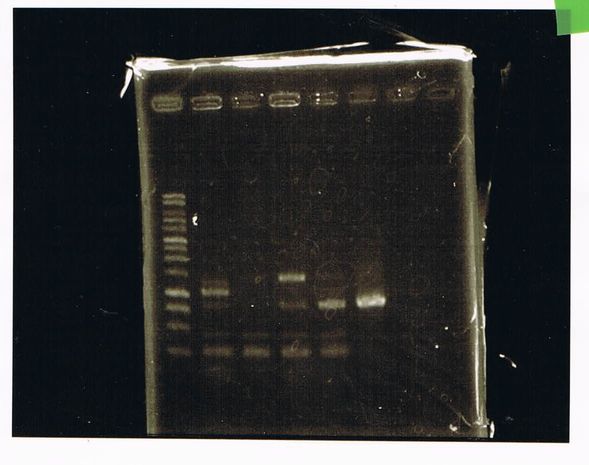
| Buffer | LacP(28.7ng/μL) | Dpn1 | MilliQ | Total |
|---|---|---|---|---|
| 2 | 10 | 0.5 | 7.5 | 20 |
| 2 | 10 | 0 | 5 | 17 |
February 23
Colony PCR
TatABCD(2 samples)
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
Predenature 94℃ 1min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 3min
→25cycles
Electrophoresis
1. 1kb ladder 2μL
2. TatABCD 1 5μL + 6×Loading Buffer 1μL
3. TatABCD 2 5μL + 6×Loading Buffer 1μL
4. 1kb ladder 2μL
Miniprep
pSB4K5 and pSB3C5
deluted it to 25 times and then measured it
pSB4K5 1 : 60.0 μg/ml 1.67(260/280) 1.98(260/230)
pSB4K5 2 : 55.0 μg/ml 1.49(260/280) 1.62(260/230)
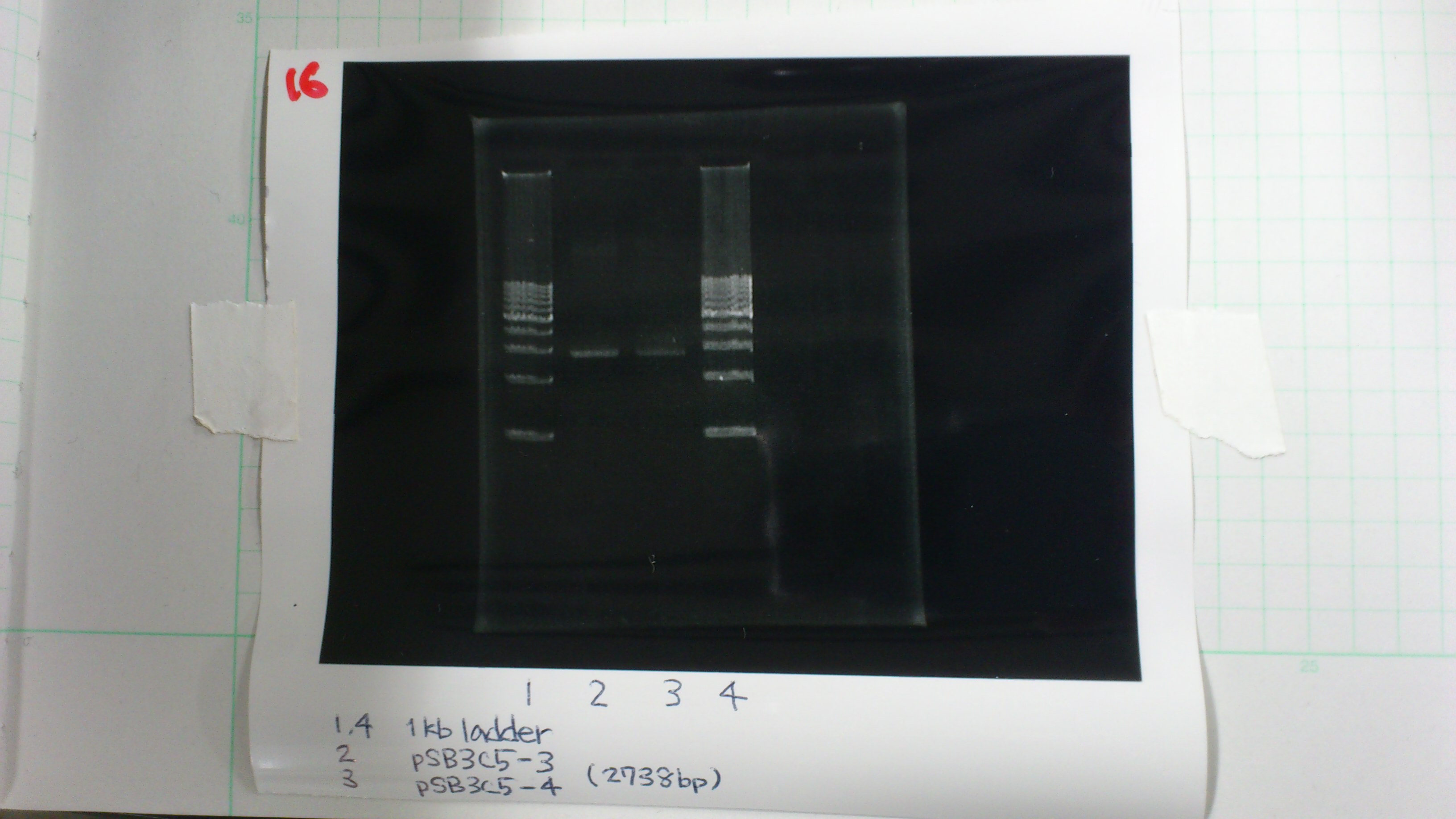
pSB3C5-3 : 3.6 μg/ml 1.57(260/280) 3.00(260/230)
pSB3C5-4 : 1.3 μg/ml 1.44(260/280) 1.04(260/230)
Liquid culture
pSB3C5-3,4
Electrophoresis
1. 1kb ladder 2μL
2. pSB3C5-3 5μL, 6×loading dye 1μL
3. pSB3C5-4 5μL, 6×loading dye 1μL
4. 1kb ladder 2μL
February 27
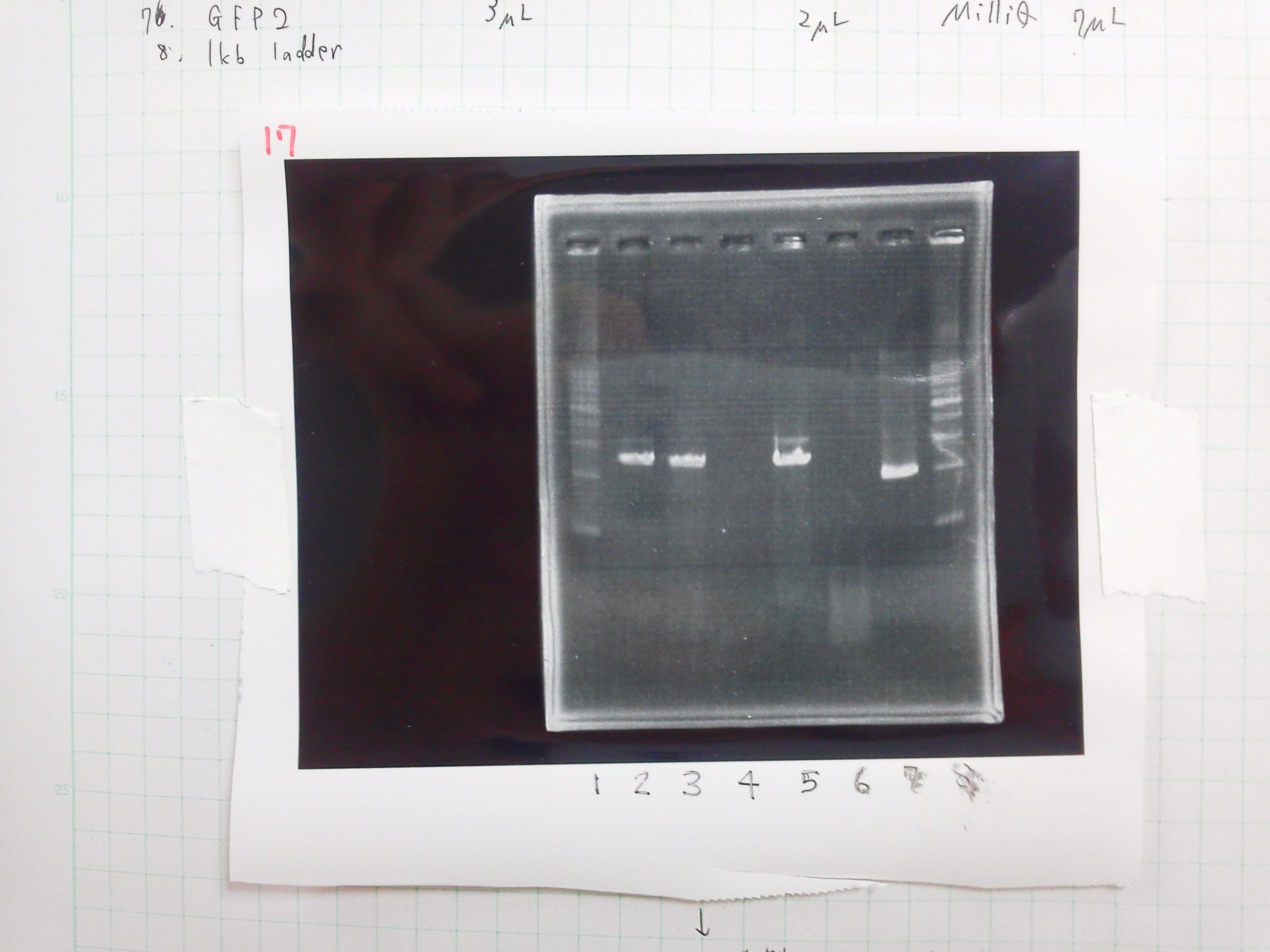
Test of Dpn1
| Buffer2 | GFP2 | BSA | MilliQ | Dpn1 |
|---|---|---|---|---|
| 3 | 3 | 0.3 | 23 | 1 |
Colony PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | MilliQ | KOD Plus | Total | |
|---|---|---|---|---|---|---|---|---|
| Colony PCR(2 samples) | 5 | 5 | 3 | 1.5 | 1.5 | 33 | 1 | 50 |
| Negative control | 5 | 5 | 3 | 1.5 | 1.5 | 34 | 0 | 50 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 59℃ 30sec
Extension 68℃ 3min
→25cycles
Electrophoresis
| lane | DNA | sample | Loading Dye | MilliQ |
|---|---|---|---|---|
| 1 | 1kb ladder | 2 | 0 | 0 |
| 2 | product of PCR1 | 5 | 1 | 0 |
| 3 | product of PCR2 | 5 | 1 | 0 |
| 4 | product of PCR(Negative control) | 5 | 1 | 0 |
| 5 | product of PCR(2/23) | 5 | 1 | 0 |
| 6 | GFP2(DPN1) | 10 | 2 | 0 |
| 7 | GFP2 | 3 | 2 | 7 |
| 8 | 1kb ladder | 2 | 0 | 0 |
Results of liquid culture
We measure this after dilute it to 10 times.
| pSB3C5-5 | pSB3C5-6 | pSB3C5-5(1% glucose) | pSB3C5-6(1% glucose) |
|---|---|---|---|
| 8.5[µg/ml] | -1.8 | -17.9 | -18.2 |
PCR
| buffer | dNTPs | MgSO4 | Primer-f(prefix) | Primer-r(suffix) | PCR purification product(1ng/µL) | MilliQ | KOD Plus | Total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 3 | 1.5 | 1.5 | 0.2 | 32.8 | 1 | 50 |
| 2 | 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 32.5 | 1 | 50 |
- PCR purification product was that purification product(75ng/µL) of electrophoresis-3 deluted to 1ng/µL
Predenature 94℃,2min
Denature 98℃,10sec
Annealing 59℃,30sec
Extension 68℃,3min
→25cycles
February 28
Electrophoresis
1. 1kb ladder
2. PCR1 →Product of gel extraction : TatABCD with prefix and suffix 105[ng/µL]
3. PCR2
Restrictive Digestion
| Buffer2 | plasmid(?) | enzyme | MilliQ | Total |
|---|---|---|---|---|
| 2 | 2 | 0.2 | 15.8 | 20 |
incubate 1 hour at 37℃
Electrophoresis
1. 1kb ladder
2. Control (without enzymes)
3. EcoR1
4. Xba1 (crystallized)
5. Xba1 (with seal)
6. Spe1
7. Pst1
8. 1kb ladder
PCR and Electrophoresis
| Quick Taq Dye Mix | primer-f | primer-r | template | MilliQ | Total |
|---|---|---|---|---|---|
| 25 | 1.0 | 1.0 | 0.5 | 22.5 | 50 |
Predenature 94℃,2min
Denature 94℃,30sec
Annealing 59℃,30sec
Extension 68℃,3min
→25cycles
Restrictive Digestion
| BufferH | TatABCD | EcoR1 | Spe1 | MilliQ | Total |
|---|---|---|---|---|---|
| 2 | 5 | 0.2 | 0.2 | 12.6 | 20 |
PCR purification
We eluted the product for 30µL MilliQ
Ligation
| Insert(TatABCD) | Vector(pSB1C3) | Ligation High | Total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
4℃, overnight
February 29
Transformation
| TatABCD | competent cell | Total |
|---|---|---|
| 1 | 10 | 11 |
Checking Restriction enzyme
| plasmid seems to be 1-18C promoter | Enzyme | Buffer | MilliQ | Total |
|---|---|---|---|---|
| 2 | 0.2 | 2 | 15.8 | 20 |
Checking TatABCD
| TatABCD | Hind3 | Buffer | MilliQ | Total |
|---|---|---|---|---|
| 5 | 0.2 | 2 | 12.8 | 20 |
PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | ColE1(6.5ng/µL) / TMAO | MilliQ | KOD Plus Neo | Total | |
|---|---|---|---|---|---|---|---|---|---|
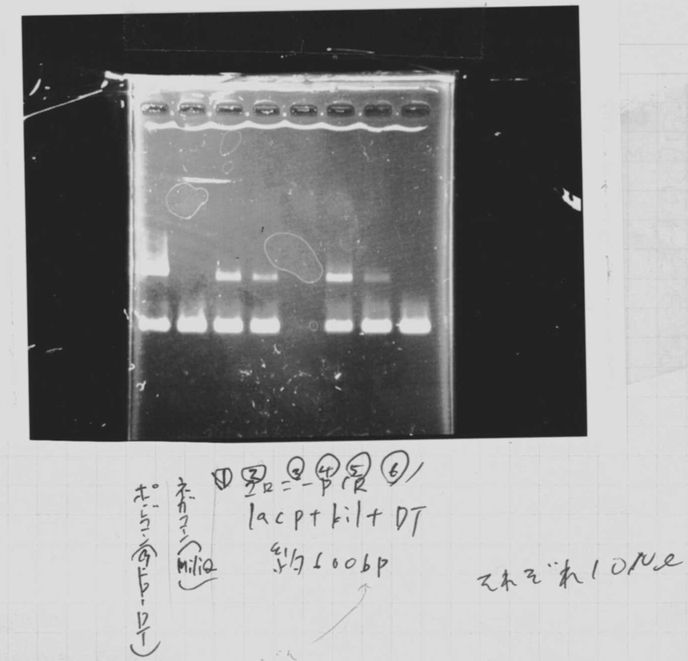
| Kil | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
| TMAO | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
Predenature 94℃,2min
Denature 98℃,10sec
Annealing 60℃,30sec
Extension 68℃,3min
→30cycles
Electrophoresis
1. 1kb ladder
2. Kil (649bp)
3. TMAO (2720bp)
4. TMAO (Quick Taq)
5. TatABCD (Quick Taq)
6. TatABCD (Hind3)
7. 1kb ladder
March 1
PCR
- TMAO
Template is gDNA and product of colony PCR gel extraction
| Buffer | gNTPs | MgSO4 | Primer-f | Primer-r | KOD Plus Neo | Template gDNA | product of gel extraction | DW | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 0 | 16 | 25 |
| 2 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0 | 2 | 14.5 | 25 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 1.5min
→25cycles
- Kil
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | colE1(6.5ng/µL) | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 0.5 | 25 |
94℃, 2min
98℃, 10sec
61℃, 30sec
68℃, 1min
→20cycles
Electrophoresis
1. 1kb ladder
2. TMAO1 (gDNA)
3. TMAO2 (product of gel extraction)
4. Kil
PCR
- kil
| Buffer | dNTPs | MgSO4 | primer-f | primer-r | Product of Purification | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
94℃, 2min
98℃, 10sec
61℃, 30sec
68℃, 30sec
20cycles
→Purification 230ng/µL
Restrictive Digestion
| Kil | EcoR1 | Spe1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 2 | 12.6 | 20 |
incubate at 37℃, for 1.5 hours
PCR Purification
Ligation
| Kil | pSB1C3 | Ligation High | Total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
at 4℃, for overnight
March 2
Ligation
| Kil | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
| TatABCD | pSB1C3 | Ligation | Total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
at 16℃ for 1 hour
Transformation
| Kil | Kil(3/1,Ligation) | TatABCD | competet cell | Total |
|---|---|---|---|---|
| 1 | 0 | 0 | 10 | 11 |
| 0 | 1 | 0 | 10 | 11 |
| 0 | 0 | 1 | 10 | 11 |
PCR
| Quick Taq | primer-r | primer-f | template | MilliQ | Total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 0.5 | 22.5 | 50 |
Electrophoresis
Restrictive Digestion
| pSB3C5-5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃, for 2 hour
Electrophoresis
1. 1kb ladder 2µL
2. pSB3C5 5µL + 6×Loading Buffer 1µL
・product of gel extraction(about 2700bp)
-30.9µg/mL
Restrictive Digestion
GFP1 ,2 ,3
| GFP | EcoR1 | Pst1 | Buffer | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃, for 2.5 hours
Restrictive Digestion
| DT | EcoR1 | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
| Constitutive Promoter | Spe1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 0.2 | 0.2 | 3 | 0.3 | 11.3 | 30 |
at 37℃, 2 hours
- J23117-1:135ng/µL, J23107-1:115ng/µL
- DT3→PCR Purification
- Promoter→Gel Extraction
Checking TMAO
| something seems to be TMAO | Buffer2 | EcoR1 | MilliQ | Total |
|---|---|---|---|---|
| 10 | 2 | 0.5 | 7.5 | 20 |
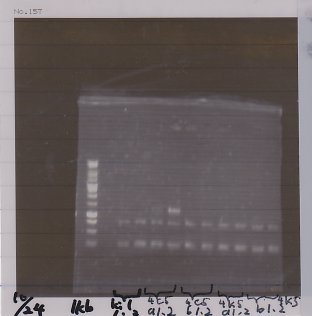
at 37℃, for 1 hour
Electrophoresis
1. 1kb ladder
2. GFP1 that had been cut by restriction enzyme
3. GFP2 that had been cut by restriction enzyme
4. GFP3 that had been cut by restriction enzyme
5. GFP1
6. GFP2
7. GFP3
8. TMAO (control)
9. TMAO (EcoR1)
10. DT (control)
11. DT (EcoR1, Xba1)
12. 1kb ladder
Checking TatABCD
| Quick Taq | primer-f | primer-r | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
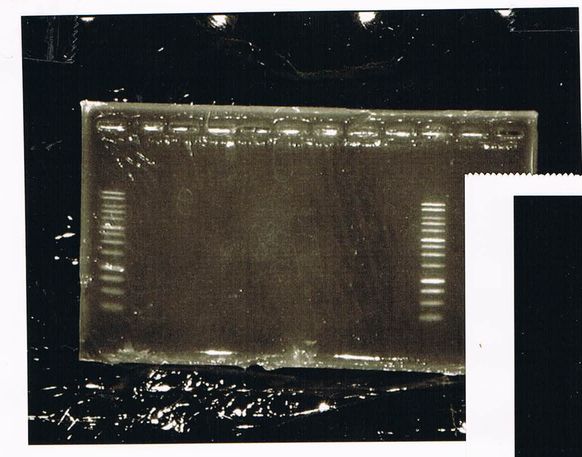
Electrophoresis
March 3
PCR
| template | buffer | dNTPs | MgSO4 | VF | VR | KOD plus | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 50 |
| 2 | 2 | 5 | 5 | 3 | 1.5 | 1.5 | 1 | 31 | 50 |
94℃, 2min
98℃, 10sec
50℃, 30sec
68℃, 1min
→30cycles
Miniprep
14.2µg/mL, 15.0µg/mL
March 4
Sequence of TatABCD
| Quick Taq | primer-f | promer-r sequence | template | MilliQ | Total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 1 | 23 | 50 |
| Quick Taq | primer-f sequence | primer-r | template | MilliQ | Total |
|---|---|---|---|---|---|
| 25 | 1 | 1 | 1 | 23 | 50 |
Colony PCR of TMAO
| buffer | dNTPs | NgSO4 | primer-f | primerr-r | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 4 | 1.5 | 1.5 | 1 | 32 | 50 |
→ethanol precipitation
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 2.5min
→25cycles
Electrophoresis
1. 1kb ladder
2. TatABCD1
3. TatABCD2
4. TMAO
5. 1kb ladder
Restrictive Digestion
| TMAO | EcoR1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 0.2 | 3 | 0.3 | 16.5 | 30 |
| TMAO | Xba1 | Pst1 | BudderM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
at 37℃ for 1 hour
Electrophoresis
Transformation
| pSB1C3 | competent cell(made at 2/8) | Total |
|---|---|---|
| 5 | 100 | 105 |
March 5
Restrictive Digestion
| pSB1C3(Xba1, Spe1) | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 0.2 | 2 | 0.2 | 7.6 | 20 |
| pSB1C3(Xba1, Spe1) | EcoR1 | BufferH | BSA | MilliQ | Total |
| 10 | 0.2 | 2 | 0.2 | 7.6 | 20 |
at 37℃ for 1 hour
→Then we did ethanol precipitation
Ligation
| Kil(EcoR1, Spe1) | pSB1C3(EcoR1) | Ligation High | Total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
at 16℃ for 1 hour
Transformation
| Kil | competent cell | Total |
|---|---|---|
| 1 | 10 | 11 |
We used commercially available competent cells in this time.
PCR
TMAO
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | Template | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 1 | 32 | 50 |
Electrophoresis
Restrictive Digestion
| LacP | pSB3C5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total | |
|---|---|---|---|---|---|---|---|---|
| 1 | 20 | 0 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| 2 | 0 | 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1 hour
1→Ethanol precipitation 45.4µg/mL
2→Gel extraction 38.7µg/mL
Ligation
| LacP | pSB3C5 | Ligation High | Total |
|---|---|---|---|
| 10 | 2 | 6 | 18 |
at 4℃ for overnight
Transformation
| LacP+pSB3C5 | competent cell | Total |
|---|---|---|
| 1 | 10 | 11 |
on ice for 30 mins.
heat shock at 42℃ for 60secs
on ice for 2 mins.
After we incubate with 200µL of SOC culture for 1 hour, we did plating on LB culture with CP
Restrictive Digestion
| GFP Plasmid | EcoR1 | Spe1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃ for 2 hours
Electrophoresis
1. Ladder 2µL
2. GFP Plasmid (already restricted) 2µL + Loading Dye 2µL + MilliQ 8µL
3. Ladder 2µL
PCR
torA signal and pspA
We did Colony PCR to pspA
| Buffer | dNTPs | MgSO4 | Primer-f | Primer-r | template(TMAO) | KOD plus | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 0.5 | 16 | 25 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
electrophoresis
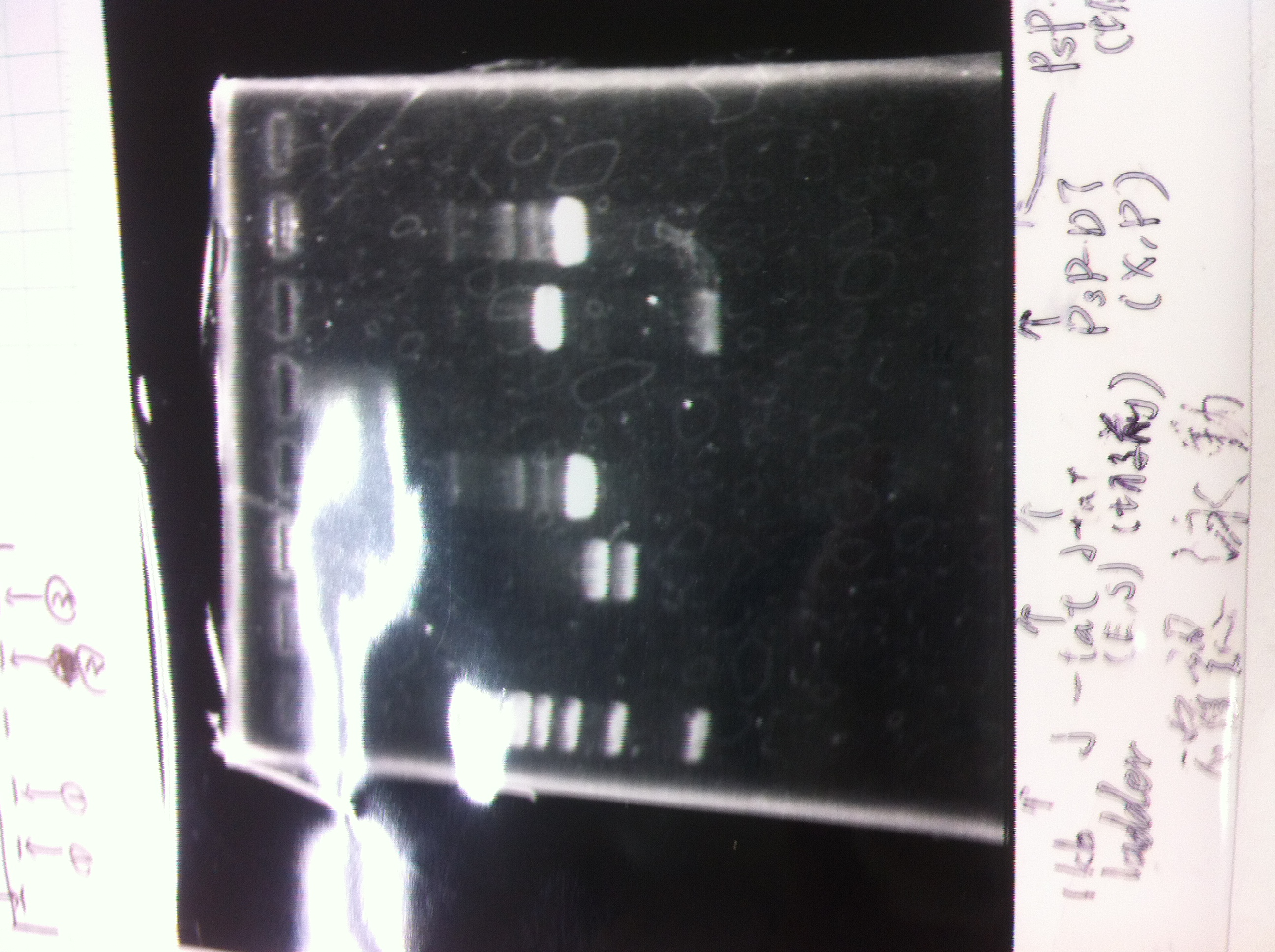
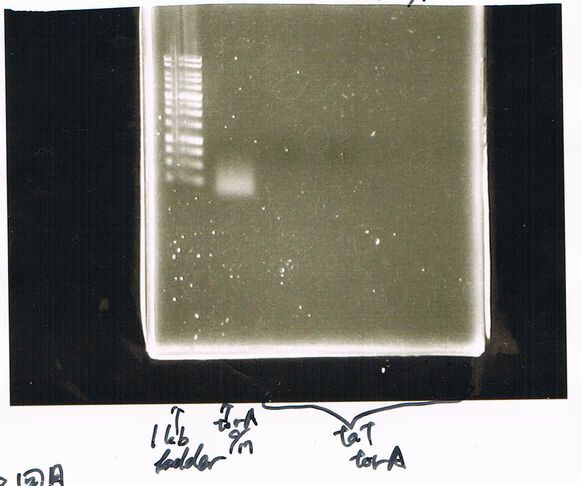
1. 100bp Ladder
2. torA signal (272bp)
3. pspA (969bp)
4. 100bp Ladder
March 6
Restrictive Digestion
| GFP | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 12 | 0.5 | 0.5 | 3 | 0.5 | 13.5 | 30 |
We did incubate at 37℃ for 1.5hours.
And we did gel extraction on 3/7 and get 40.0μg/mL GFP.
PCR
| buffer | dNTPs | MgSO4 | Primer-f | Primer-r | template | KOD plus neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 1 | 32.5 | 50 |
94℃ 2min
98℃ 10sec
60℃ 30sec
68℃ (torA 10sec / pspA 30sec) 25 cycles
We used product of PCR on 3/5 of torA and pspA as template.
Restrictive Digestion
| Kil | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 3 | 0.3 | 16.1 | 30 |
| DT | EcoR1 | Xba1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃ for 2 hours
→ purification 37.7ng/μL
Ligation
| Kil | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
- Kil : 350fmol
- pSB1C3 : 29fmol
at 16℃ for overnight
March 7
Electrophoresis
1. 1kb ladder 2µL
2. pspA (PCR product)2.5µL + Loading Dye 0.5µL
3. GFP 30µL + Loading Dye 6µL
4. 1kb ladder 2µL
- The GFP was gel extracted on 3/6 and concentrated by Vacuum in 150µg/mL
Ligation
| VectorDNA | GFP | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 15 | 10 | 30 |
Restrictive Digestion
| torA | EcoR1 | Spe1 | bufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 3 | 0.3 | 16.1 | 30 |
| pspA | EcoR1 | Spe1 | bufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.3 | 0.3 | 3 | 0.3 | 21.1 | 30 |
at 37℃ for 1.5 hours
Purification
torA→31.8ng/µL
pspA→49.3ng/µL
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 3 | 3 | 3 | 9 |
| pspA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 4℃, for overnight
- torA→31.8ng/µL×3µL=95.4ng=0.529pmol
- pSB1C3→19.4ng/µL×3µL=58.2ng=0.042pmol
- pspA→49.3ng/µL×4µL=197.2ng=0.308pmol
- pSB1C3→19.4ng/µL×2µL=38.8ng=0.029pmol
March 8
Restrictive Digestion
| pSB4K5 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 3 | 0.2 | 6.4 | 30 |
at 37℃ for 1 hour.
→Purification : 36.6ng/µL
Ligation
| Kil | pSB4K5 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
at 4℃ for overnight
- Kil→37.7ng/µL×10µL=377ng=879fmol
- pSB4K5→36.6ng/µL×1µL=36.6ng=86fmol
Liquid culture
LacP + pSB3C5 -1, 2
Transformation
| torA | pspA | competent cell | Total |
|---|---|---|---|
| 1 | 0 | 10 | 11 |
| 0 | 1 | 10 | 11 |
We use commercially available competent cells in this time.
March 9
Restrictive Digestion
| TatABCD | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 3 | 0.3 | 16.3 | 30 |
at 37℃ for 1hour
→purification : 75.0μg/mL (1.11 260/280 , 0.81 260/230)
Miniprep
LacP + pSB3C5 1 80.5μg/mL (1.78 260/280 , 2.00 260/230)
LacP + pSB3C5 2 107.2μg/mL (1.83 260/280 , 1.90 260/230)
Colony PCR
| Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 6sec
25cycles
Ligation
| TatABCD | constP J23107 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
TatABCD : 227fmol
constP J23107 : 21fmol
Transformation
| pspA | torA | Kil | competent cell | |
|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 10 |
| 2 | 0 | 1 | 0 | 10 |
| 3 | 0 | 0 | 1 | 10 |
Miniprep
4mL of plusgrow which had been cultured for overnight.
pSB4K5 : 80.5μg/mL
March 10
Screening PCR
| Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Then we did electrophoresis to confirm.
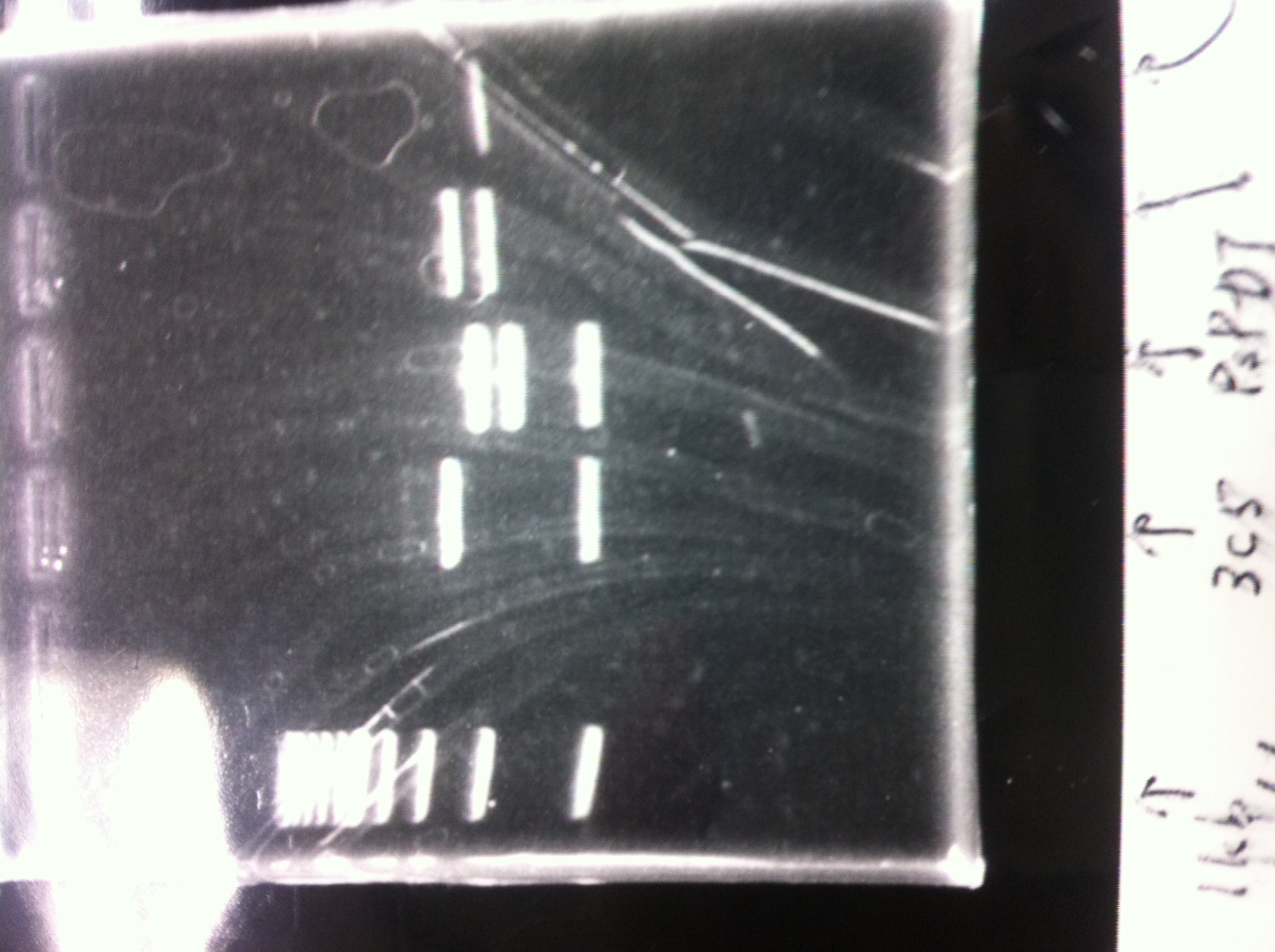
1. 1kb ladder
2. kil (649bp)
3. 4. 5. pspA (969bp)
6. 1kb ladder
1. 100bp ladder
2. 3. 4. torA signal
Restrictive Digestion
| LacP-pSB3C5 | Spe1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 29.0ng/μL
| torA | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 2 | 0.2 | 7.4 | 20 |
for 2.5 hours at 37℃
→purification 91.8ng/μL
Ligation
| torA | LacP-pSB3C5 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
torA : 929fmol
LacP-pSB3C5 : 284fmol
for overnight at 4℃
March 11
Miniprep
We used 3μL of plus grow that we had cultured for overnight.
torA : 62.8ng/μL
Screening PCR
| Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
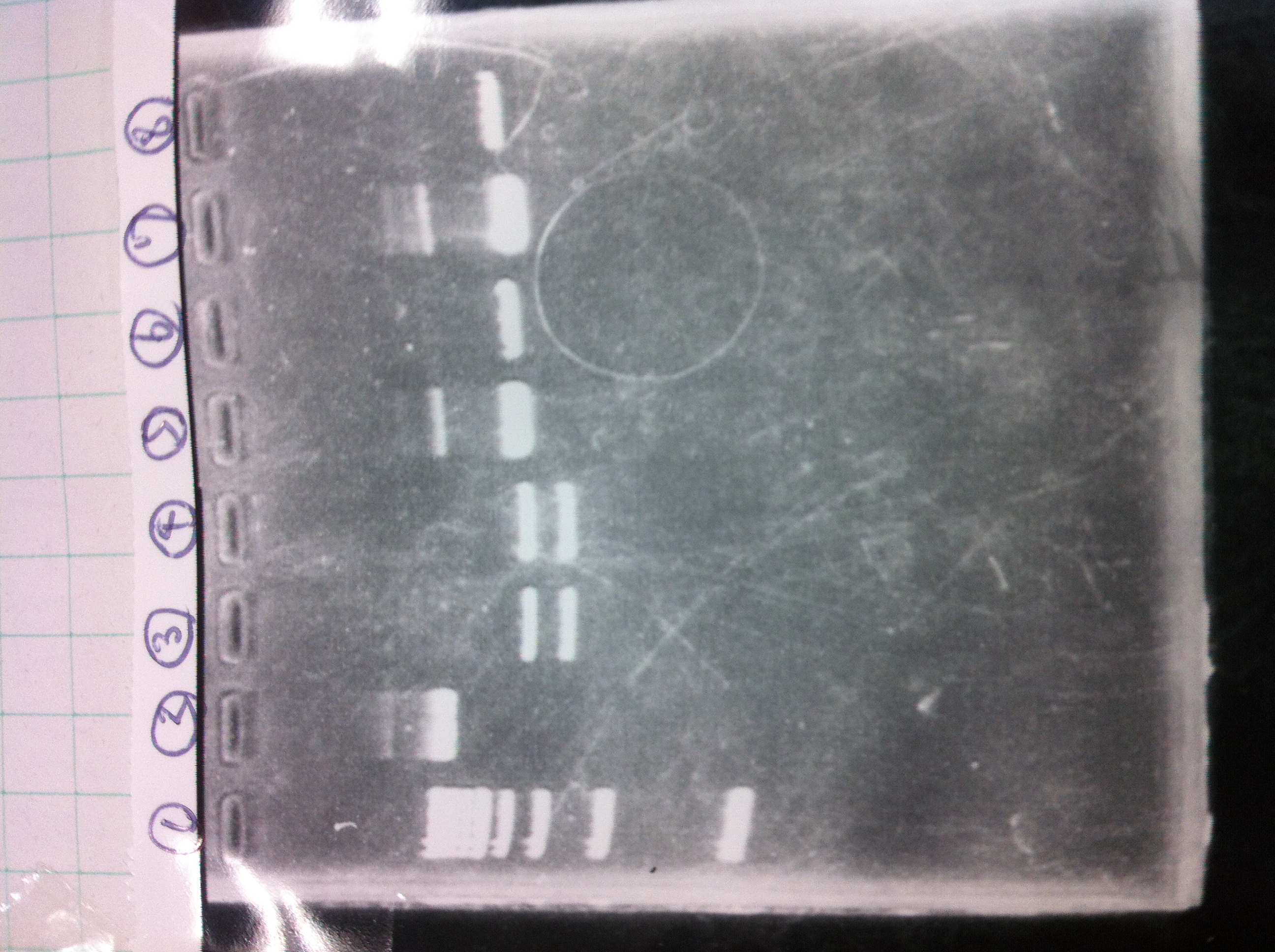
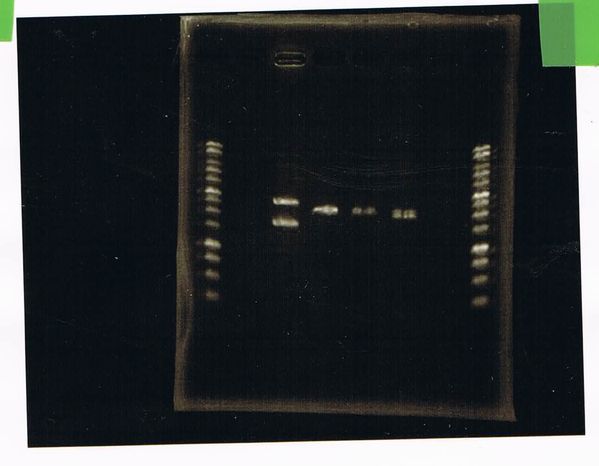
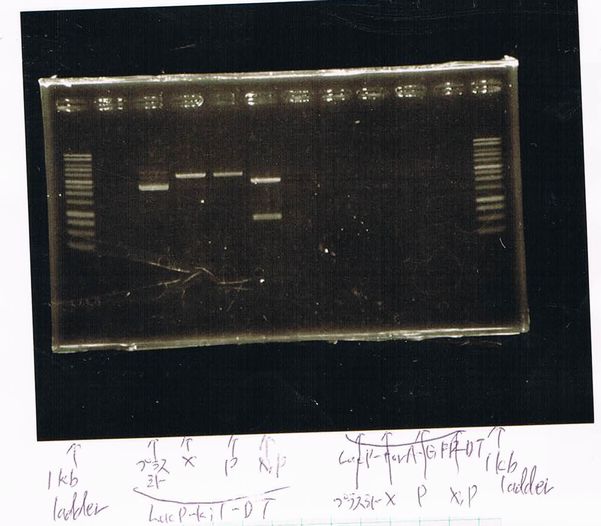
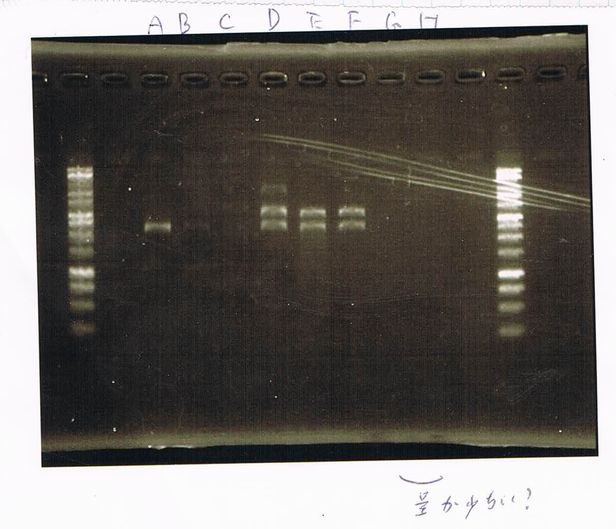
Electrophoresis
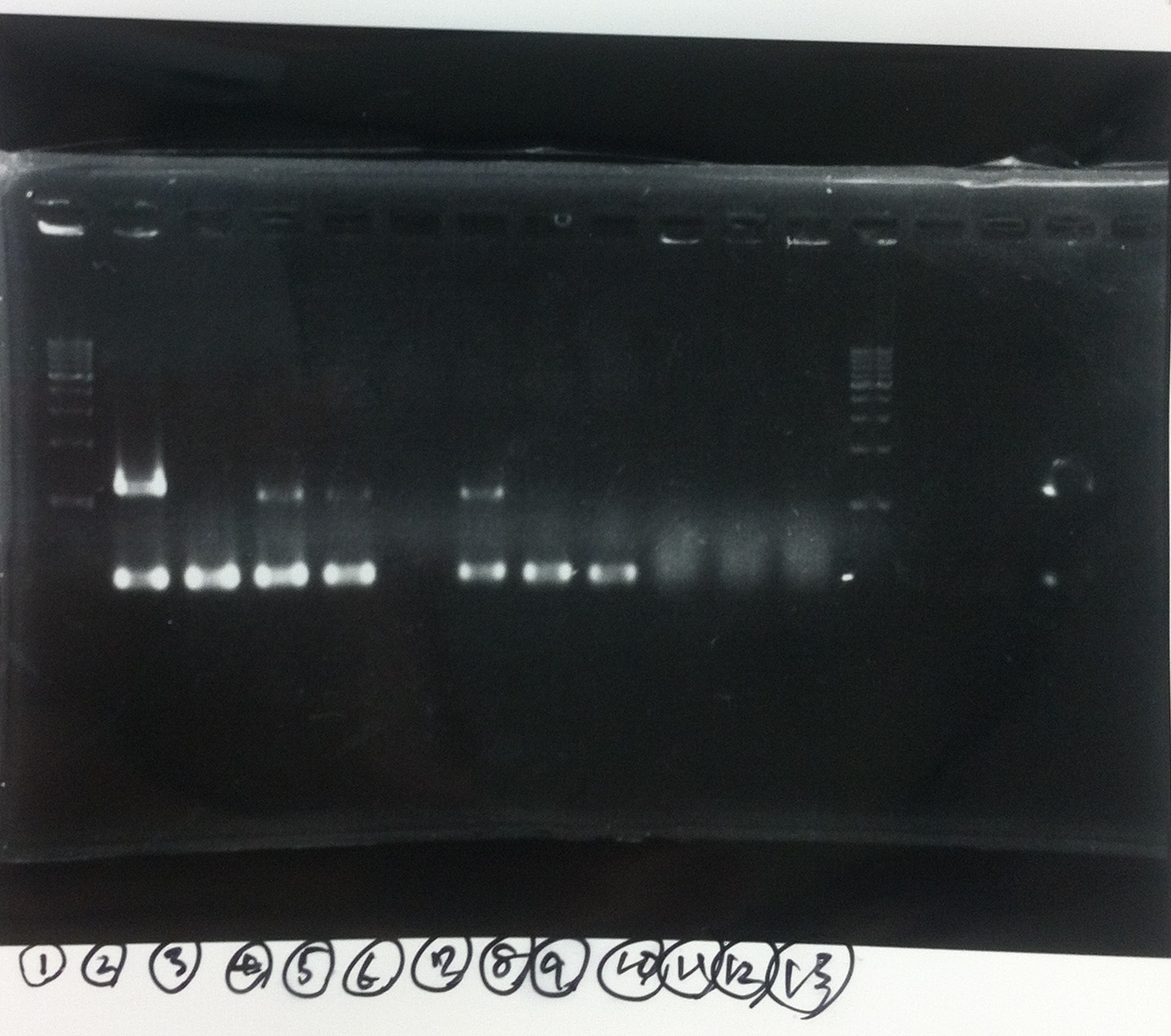
1. 1kb ladder
2〜11. pspA (969bp)
12. 1kb ladder
The results were shown as photograph in the right.
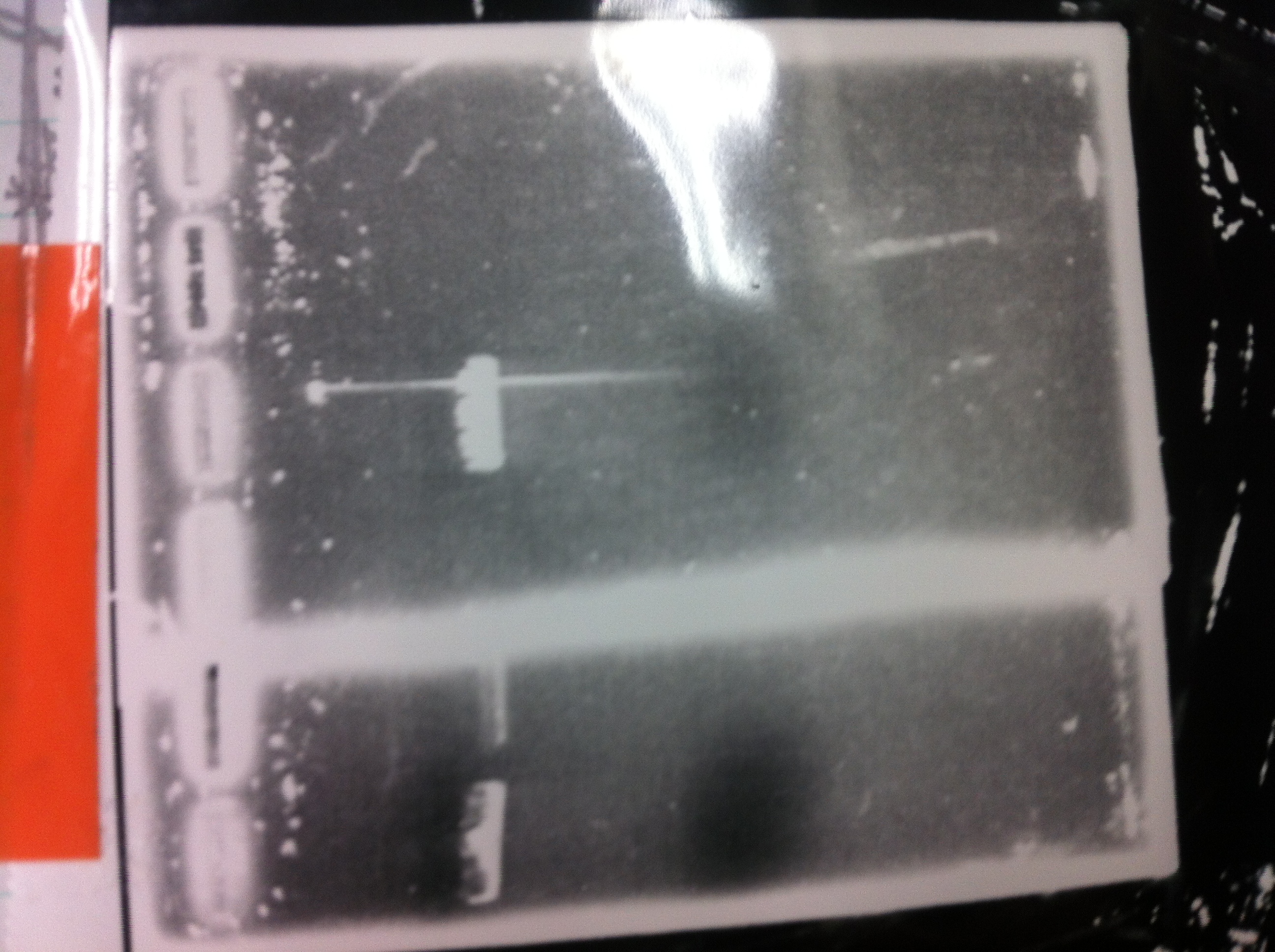
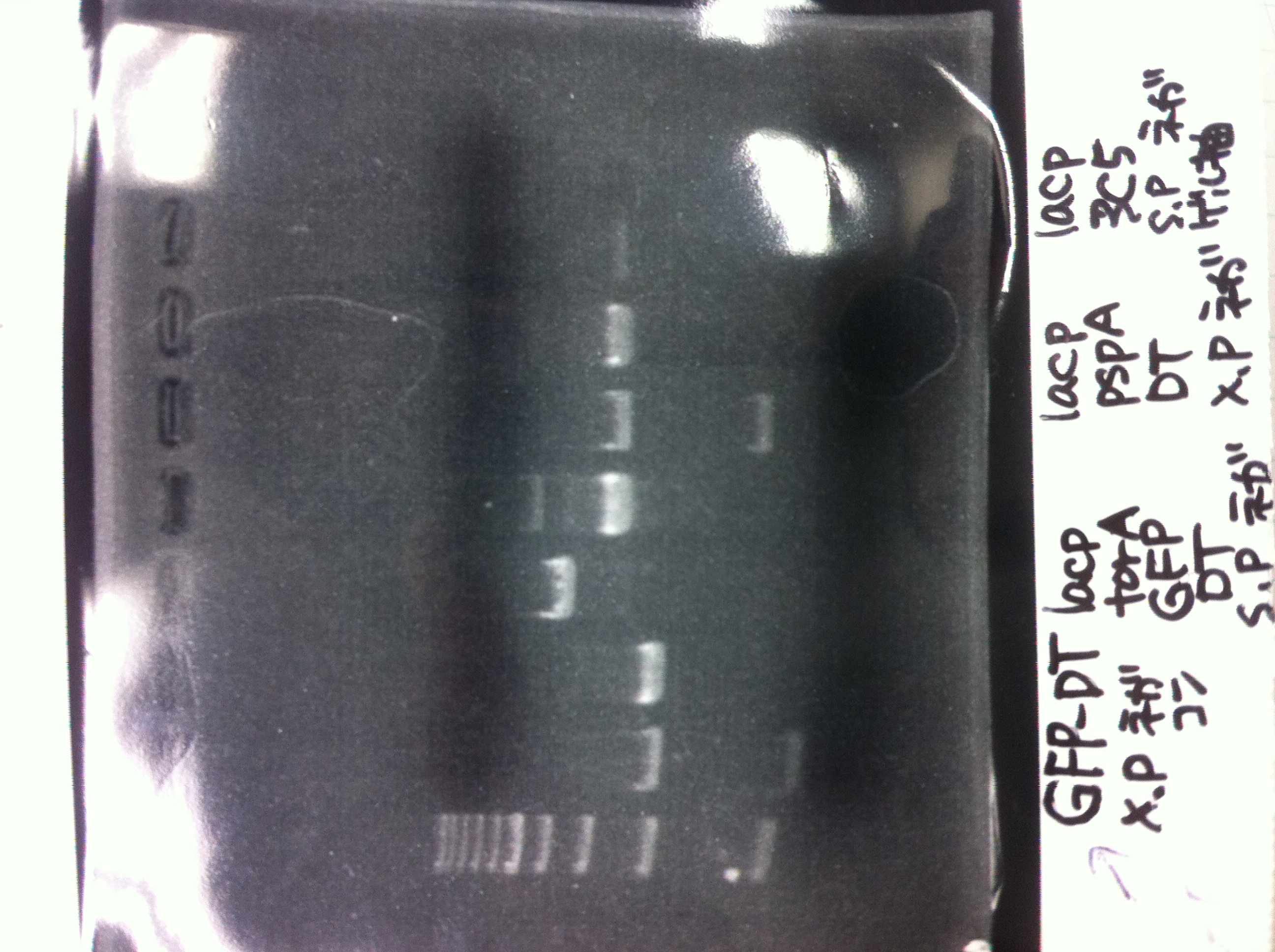
It seemed that there were shorter sample than expected sample, so we did electrophoresis with pspA which was product of PCR and pspA which had already cut with EcoR1 and Spe1.
1.1kb ladder
2. pspA (PCR product)
3, pspA (Eco, Spe)
4〜6, pspA (colony PCR)
7,1kb ladder
The results were shown as photograph in the right.
March 12
Transformation
| DT(1ng/μL) | DT(0.1ng/μL) | Kil | LacP-torA | MilliQ | competent cell | Total |
|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 20 | 21 |
| 0 | 1 | 0 | 0 | 0 | 20 | 21 |
| 0 | 0 | 5 | 0 | 0 | 50 | 51 |
| 0 | 0 | 0 | 5 | 0 | 50 | 51 |
| 0 | 0 | 0 | 0 | 1 | 20 | 21 |
Restrictive Digestion
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.3 | 15.7 | 30 |
at 37℃ for 4 hours
→ We did purification and got 48.3ng/μL pspA.
Ligation
| pspA | pSB1C3 | MilliQ | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 4 | 2 | 0 | 3 | 9 |
| 2 | 2 | 0 | 2 | 6 |
| 0 | 2 | 2 | 2 | 6 |
March 13
Miniprep
pSB1C3 74.2µg/mL 1.65 (260/280) 1.31 (260/230)
March 14
Restrictive Digestion
| pSB1C3 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 4 | 0.4 | 15.2 | 40 |
We did gel extraction and got 47.2ng/µL pSB1C3 but we did not cut out RFP.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | Total |
| 4 | 2 | 3 | 9 |
at 16℃, for 1 hour
- torA : 0.707pmol
- pSB1C3 : 0.068pmol
Liquid culture
We cultured LacP-pSB3C5 1,2,3,4,5 (CP tolerance)on culture with Amp.
→Only 4 which did not be cultured succeeded.
March 15
Liquid culture
We cultured LacP-pSB3C5 6,7,8,9,10 on culture with ampicillin.
→6,8,9,10 were succeeded.
Restrictive Digestion
| GFP | EcoR1 | Spe1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
| DT | EcoR1 | Xba1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
for 2 hours at 37℃.
Miniprep
pspA (pSB1C3) 40.5ng/µL
Ligation
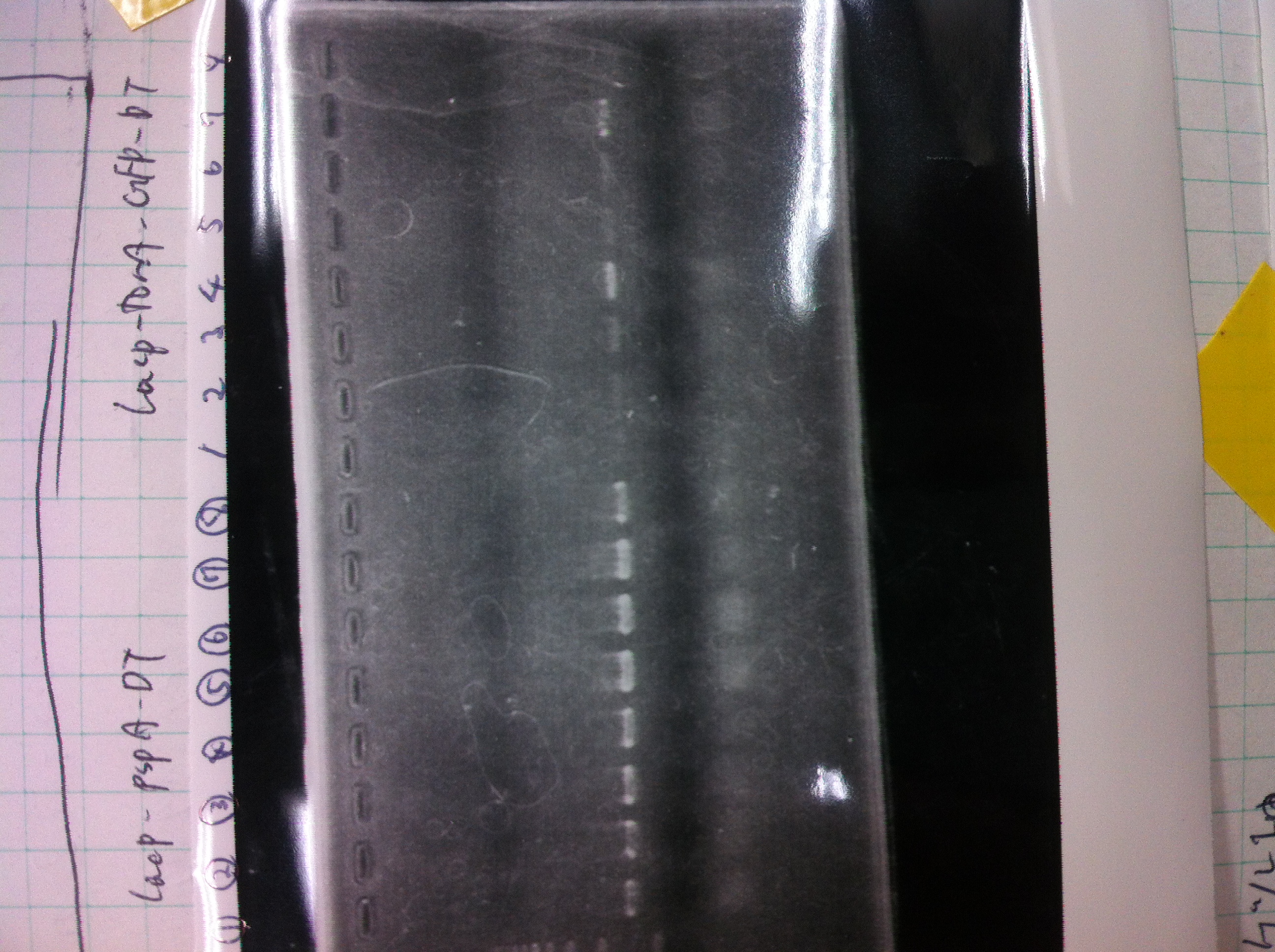
| pspA | DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 1.5 | 3 | 9.5 |
- pspA : 385fmol
- DT : 36fmol
Transformation
| J23107-TatABCD | DT (0.1ng/µL) | DT (0.01ng/µL) | pspA-DT | competent cells on 3/15 | Total |
|---|---|---|---|---|---|
| 2 | 0 | 0 | 0 | 20 | 22 |
| 0 | 2 | 0 | 0 | 20 | 22 |
| 0 | 0 | 2 | 0 | 20 | 22 |
| 0 | 0 | 0 | 2 | 20 | 22 |
Screening PCR
Kil, pspA and torA
| Quick Taq | Primer-r | Primer-f | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
March 16
Miniprep
torA (pSB1C3) 68.8ng/µL
Kil (pSB4K5) 92.7ng/µL
torA was red for some reason. We do not know why.
Colony PCR
| Quick Taq | Primer-r | Primer-f | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 6sec
→25cycles
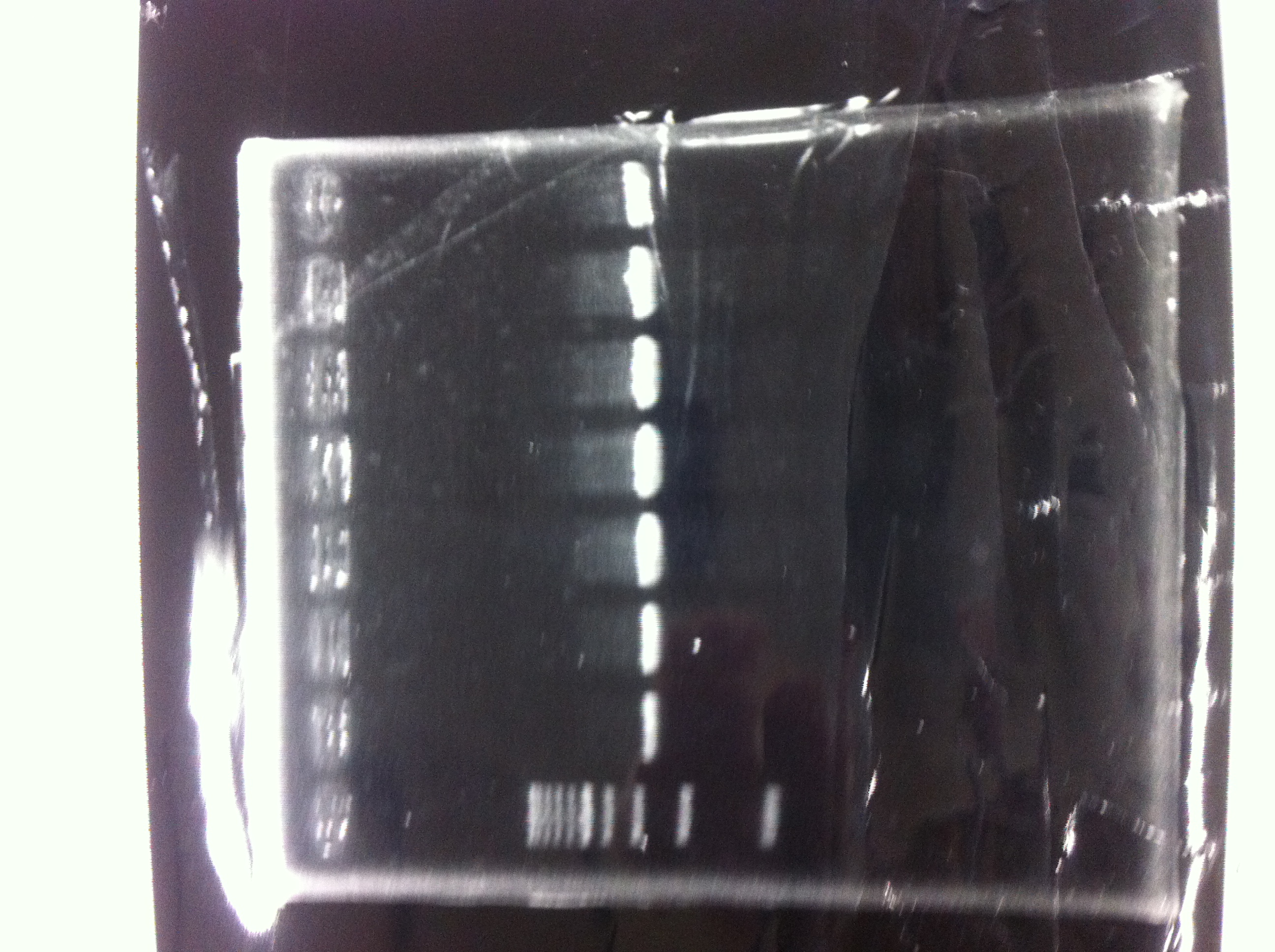
Electrophoresis
The results were shown as photograph in the right.
Checking Transformation Efficiency
competent cells that were made on March 15.
DNA : 0.02ng → 668 colonies Transformation Efficiency : 3.3×10^7
DNA : 0.2ng → 1739 colonies Transformation Efficiency : 8.7×10^6
Restrictive Digestion
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | BufferH | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 20 | 0.2 | 0.2 | 0.3 | 3 | 0 | 6.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 0 | 2 | 12.6 | 20 |
at 37℃, for 2 hours.
We did gel extraction for product with EcoR1, Spe1. We got 46.1ng/µL pSB1C3.
| Kil(pSB4K5) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
| pspA (pSB1C3) | EcoR1 | Pst1 | BSA | BufferH | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
for overnight at 37℃.
We did this to confirm.
Ligation
| torA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
| MilliQ | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 2 | 3 | 9 |
at 16℃, for overnight
- torA→767fmol
- pSB1C3→68fmol
March 17
Miniprep
J23107-TatABCD 72.7ng/µL
pspA-DT 50.5ng/µL
Checking the Insert
| J21037-TatABCD | EcoR1 | Pst1 | BSA | BufferH | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Success.
| pspA-DT | EcoR1 | Pst1 | BSA | BufferH | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
Failed.
March 19
Restrictive Digestion
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did Gel extraction and got 17.2ng/µL of DT.
Ligation
| Kil | DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 2 | 6 | 18 |
We did this for an hour at 16℃.
Restrictive Digestion
| GFP | EcoR1 | Spe1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 0.5 | 3 | 15.5 | 30 |
We did this for 4 hours at 37℃
Ligation
| pspA | DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 5 | 5 | 15 |
| pspA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 1 | 3 | 8 |
We did these for an hour at 16℃.
- pspA (5µL)→377fmol
- DT→39fmol
- pspA (4µL)→339fmol
- pSB1C3→34fmol
Transformation
pspA-DT, pspA (pSB1C3), torA (pSB1C3) and GFP-DT
March 20
Screaning PCR
| Quick Taq | Primer-R | Primer-F | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→○
- GFP-DT→○
- torA→×
- Kil-DT 6 of 8 sumples→○
| Quick Taq | VR | VF | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
- pspA→○
- pspA-DT→×
Restrictive Digestion
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.3 | 0.3 | 0.3 | 3 | 16.1 | 30 |
| DT | EcoR1 | Xba1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for overnight at 37℃. And we did purification.
torA 34.2ng/µL
DT 28.0ng/µL
March 21
Miniprep
GFP-DT-1 : 77.7ng/µL
GFP-DT-2 : 67.6ng/µL
Restrictive Digestion
| pSB1C3 | EcoR1 | Spe1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
| 5 | 0.2 | 0 | 0.2 | 2 | 12.6 | 20 |
We did this for 3 hours at 37℃, and then we did gel extraction. We got 25.1ng/µL pSB1C3
| GFP-DT | EcoR1 | Pst1 | Xba1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 0 | 0.2 | 2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 0.2 | 2 | 12.6 | 20 |
| 10 | 0.2 | 0 | 0.2 | 0.3 | 3 | 16.3 | 30 |
We did this for 3 hours at 37℃, and then we did Purification. We got 30.7ng/µL GFP-DT.
Ligation
| torA | pSB1C3 | pspA | DT | GFP-DT | Ligation High Ver.2 | Total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 1 | 0 | 0 | 0 | 3 | 8 |
| 2 | 0 | 1 | 7 | 0 | 0 | 4 | 12 |
| 3 | 0 | 0 | 5 | 3 | 0 | 4 | 12 |
| 4 | 3 | 0 | 0 | 0 | 5 | 4 | 12 |
We did this for an hour at16℃.
March 22
PCR
We did PCR to amplify torA_signal that was product of PCR at March 5 with redesigned primers.
| Buffer | dNTPs | MgSO4 | Primer-F | Primer-R | Template | MilliQ | KOD plus neo | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 0.5 | 32.5 | 1 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 10sec
→30cycles
→Purification 110.7ng/µL
Restrictive Digestion
| torA | EcoR1 | Spe1 | BSA | BufferM | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.2 | 0.2 | 0.3 | 3 | 16.3 | 30 |
Ligation
| torA | pSSB1C3 | pspA | DT | GFP-DT | Ligation high | Total | |
|---|---|---|---|---|---|---|---|
| 1 | 4 | 3 | 0 | 0 | 0 | 4 | 11 |
| 2 | 3 | 0 | 0 | 0 | 3 | 3 | 9 |
| 3 | 0 | 0 | 5 | 5 | 0 | 5 | 15 |
- torA (4µL)→512fmol
- pSB1C3→54fmol
- torA (3µL)→384fmol
- GFP-DT→36fmol
- pspA→377fmol
- DT→65fmol
March 23
Screening PCR
torA (pSB1C3), torA-GFP-DT and pspA-DT
| Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
E.coli in liquid culture that had pspA(pSB1C3) also expressed RFP.
Restrictive Digestion
| pspA | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.3 | 0.3 | 3 | 0.3 | 21.1 | 30 |
And then we did ethanol precipitation
Ethanol precipitation
pspA 11.5ng/µL.
Miniprep
LacP+pSB3C5-8 77.9µg/mL
LacP+pSB3C5-10 69.0µg/mL
Kil+DT-4 58.0µg/mL
Kil+DT-7 15.2µg/mL
Restrictive Digestion
| LacP+pSB3C5-8 | Spe1 | Pst1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| Kil+DT-4 | Xba1 | Pst1 | Buffer2 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1.5 hours
And then we did Gel extraction.
Gel Extraction
LacP+pSB3C5-8 40.4µg/mL
Kil+DT-4 26.8µg/mL
March 26
Miniprep
torA(pSB1C3) 18.5[ng/µL]
torA-GFP-DT 20.0[ng/µL]
Restrictive Digestion
| torA(pSB1C3) | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.2 | 0.2 | 2 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 2 | 0.2 | 12.6 | 20 |
| torA-GFP-DT | EcoR1 | Xba1 | Pst1 | BufferH | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 0.2 | 0 | 0.2 | 2 | 0 | 0.2 | 12.4 | 20 |
| 5 | 0.2 | 0 | 0 | 2 | 0 | 0.2 | 12.6 | 20 |
| 20 | 0 | 0.2 | 0.2 | 0 | 3 | 0.3 | 6.3 | 30 |
We did Gel extraction and then got ??? 28.7[ng/µL]
Ligation
| LacP (pSB3C5) | torA-GFP-DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
- LacP : 22fmol
- torA-GFP-DT : 197fmol
| pspA | pSB1C3 | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
| pspA | DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 1 | 5 | 16 |
- pspA : 180fmol
- pSB1C3 : 18fmol
- DT : 16fmol
| LacP(pSB3C5) | Kil-DT | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
for 2 hours at 16℃
Transformation
| LacP-Kil-DT | competent cell | Total |
|---|---|---|
| 1 | 10 | 11 |
March 27
Miniprep
We retryed miniprep of torA(pSB1C3).
We got torA and its concentration was 39.3[ng/µL].
Transformation
| Name | Well | Sample | Competent Cells | Total | Plate | Colony |
|---|---|---|---|---|---|---|
| BBa_K117004 | 14J(2011 plate2) | 5 | 20 | ? | ? | ? |
We added 100[µL] of culture medium before we started culturing the E.coli.
Screening PCR
| Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
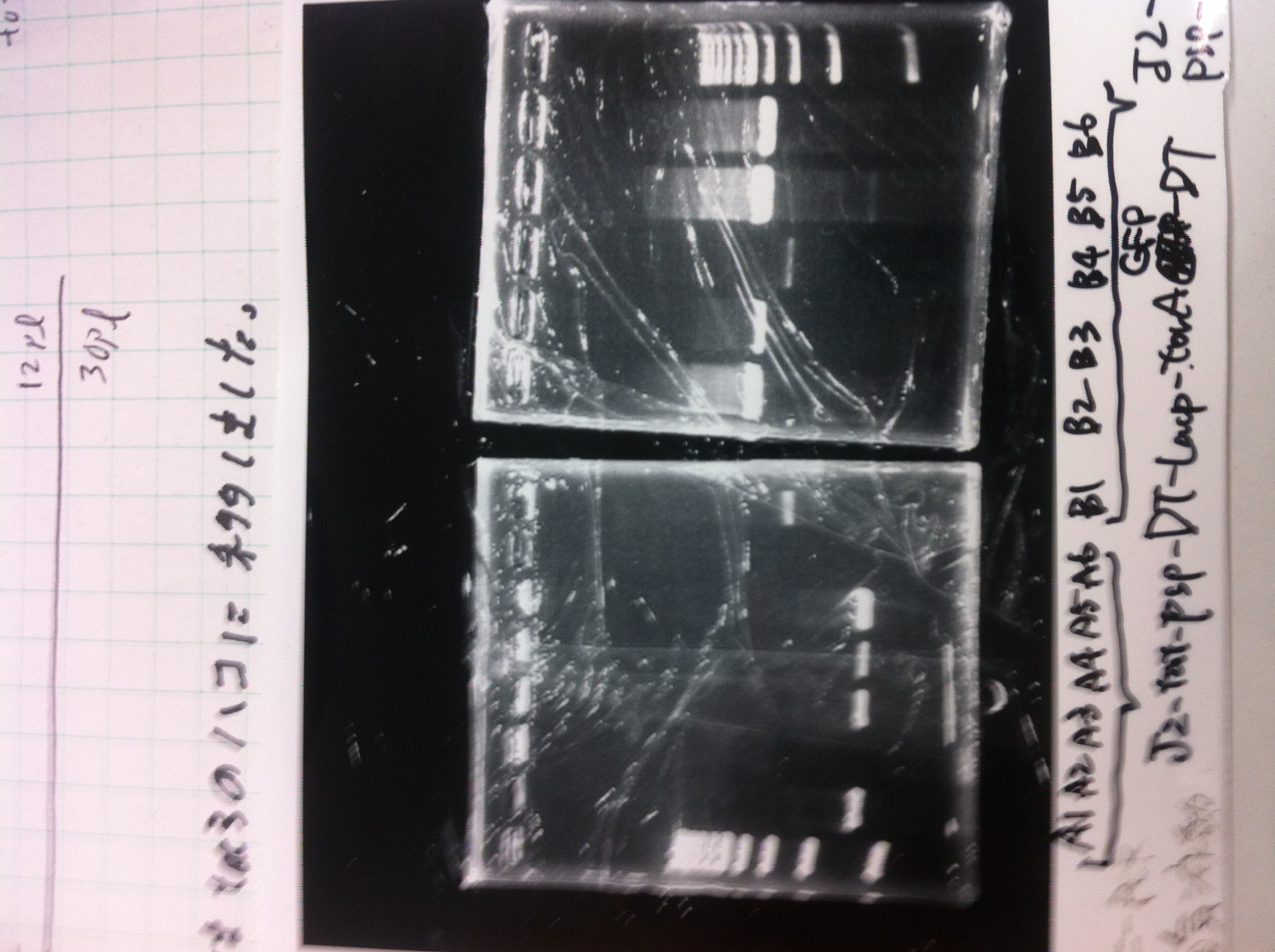
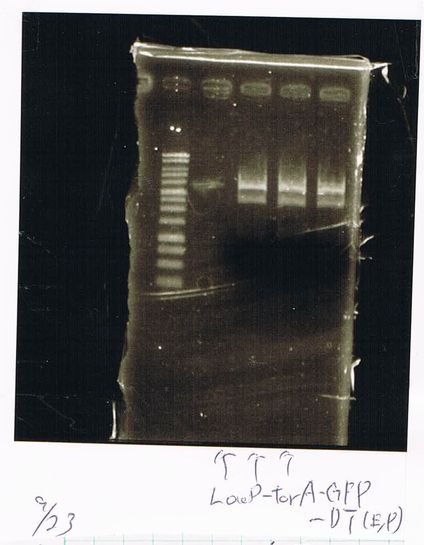
LacP-torA-GFP-DT 1, 3, 5, 6, 8, 9 was successful.
pspA-DT was failed.
E.coli that had pspA(pSB1C3) did not make any colony.
LacP-Kil-DT 1,2,3,4,5,6,7,8 was failed.
Liquid culture
LacP-torA-GFP-DT
July 23
Transformation
| DT(64.4ng/μl) | Competent cell | Plating in SOC medium | Total |
|---|---|---|---|
| 1 | 20 | 100 | 121 |
July 24
Plating in SOC medium
July 25
Transformation
- BBa_J23113 from iGEM Kit
- LacP from iGEM Kit
July 27
Restrictive Digestion
| Kil +DT (44.0 ng/μl) | 10xBuffer2 | BSA | Xbal | MilliQ | Total |
|---|---|---|---|---|---|
| 13.6 | 3.0 | 0.5 | 0.5 | 11.9 | 30.0 |
| LacP(28.7ng/μl) | 10xBuffer2 | BSA | Spe1 | Pst1 | Total |
|---|---|---|---|---|---|
| 20.9 | 3.0 | 0.5 | 0.5 | 4.6 | 30.0 |
July 30
Ligation
| kil+DT(Xba1,Pst1) | LacP(Spe1,Pst1) | Ligation High | Total |
|---|---|---|---|
| 30 | 6 | 36 | 72 |
July 31
Restriction Enzyme Processing
| torA-GFP+DT | 10×M Buffer | BSA | MilliQ | Xba1 | Pst1 | Total |
|---|---|---|---|---|---|---|
| 2.5 | 3.0 | 0.5 | 23.4 | 0.3 | 0.3 | 30.0 |
Liquid Culture
J23113(backbone J61002)
August 1
Restrictive Digestion
| LacP+kil+DT | BSA | 10×H Buffer | EcoR1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5.0 | 0.5 | 3.0 | 0.5 | 0.5 | 20.5 | 30.0 |
| LacP middle copy | BSA | 10×H Buffer | EcoR1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10.0 | 0.5 | 3.0 | 0.5 | 0.5 | 15.5 | 30.0 |
MIniprep
J23113(backbone J61002)
Liquid culture
J23113(backboneJ61002) Three test tubes of 4 mL LB medium with ampicillin 37℃ overnight
August 3
Restrictive Digestion
| pSB4K5 | BSA | 10×H Buffer | EcoR1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|---|
| 8.0 | 0.5 | 3.0 | 0.5 | 0.5 | 17.5 | 30.0 |
| B0034 | BSA | 10×H Buffer | Spe1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|---|
| 12.0 | 0.5 | 3.0 | 0.5 | 0.5 | 13.5 | 30.0 |
37℃ 2h
DNA purification
Ligation
| LacP+kil+DT | pSB4K5 | ligation high | Total |
|---|---|---|---|
| 15 | 15 | 15 | 45 |
Miniprep
- J23113(backbone J61002) 218.0μg/mL
- J23113(backbone J61002) 252.5μg/mL
- J23113(backbone J61002) 201.0μg/mL
- pSB3C5 45.0μg/ml 1.60 260/280 1.80 260/230
- pSB4C5 213.0μg/mL 1.77 260/280 4.40 260/230
August 10
Ligation
| B0034 Spe1 Pst1 | GFP+DT Xba1 Pst1 | ligation high | Total |
|---|---|---|---|
| 2 | 3 | 4 | 9 |
Transformation
- RBS+GFP+DT
- LacP+kil+DT
- RBS(for control)
- To put 2ng DNA in 20μL competent cell and leave it on ice for 30min
- Heat shock for 60s at 42℃
- To leave on ice for 2min
- To spread on ampicillin LB plate
August 13
Liquid culture
B0034 3mL ×3
- 37℃ overnight
August 14
Miniprep
- B0034 0μg/mL
- B0034 235.5μg/mL
- B0034 76.5μg/mL
Colony PCR
LacP+kil+DT ×5
| 2×Quick Taq | VF | VF | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
25cycles
pspA
| 10×Buffer for KOD-Plus-Ver2 | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f-p | Primer PsPA r-s | KOD-Plus- | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
August 15
Colony PCR
pspA
| 2×Quick Taq | pspA r-s | pspA f-p | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
56℃, 30sec
68℃, 1min
→35cycles
August 16
Electrophoresis
positive control(GFP+DT)
negative control(MilliQ)
① ~⑥ colony PCR LacP+kil+DT
①1kb ladder
②Positive control (GFP+DT)
③Negative control (MilliQ)
④~⑨LacP+kil+DT ①~⑥
⑩~⑫pspA 10μL ,6×buffer 2μL
⑬1kb ladder
August 17
Colony PCR
pspA
| 10×Buffer for KOD-Plus-Ver2 | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD-Plus- | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 3 | 3 | 1.5 | 1.5 | 1 | 35 | 50 |
| 5 | 3 | 5 | 1.5 | 1.5 | 1 | 33 | 50 |
negative control(MilliQ 50μL)
94℃, 2min
94℃, 15sec
55℃, 30sec
68℃, 1min
→35cycles
Electrophoresis
① 1kb ladder
②, ③ MgSO4 3μL, pspA
④, ⑤ MgSO4 5μL, pspA
⑥ negative control(MilliQ)
⑧ 1kb ladder
August 20
Restrictive Digestion
| J23113(201.0ng/μL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 0.5 | 14.5 | 30 |
August 21
Colony PCR
pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 3 | 5 | 1.5 | 1.5 | 1 | 33 | 50 |
| 5 | 3 | 7 | 1.5 | 1.5 | 1 | 31 | 50 |
| 5 | 3 | 10 | 1.5 | 1.5 | 1 | 28 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→25cycles
Restrictive Digestion
| J23113(218ng/μL) | Spe1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 0.5 | 14.5 | 30 |
| torA-GFP-DT(20ng/μL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 5 | 0.6 | 0.6 | 3 | 0.5 | 20.3 | 30 |
Electrophoresis
① 1kb ladder
②, ③ torA-GFP-DT(Xba1,Pst1)
④, ⑤ J23113(Spe1,Pst1)
Transformation
- LacP-torA-GFP-DT(Backbone pSB3C5)
- BBa_K11704(Backbone pSB1A2)
Electrophoresis
laneA-C. pspA
laneD. pspA(without primer)
Colony PCR
- pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f | Primer PsPA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
- negative control for pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer PsPA f-p | Primer PsPA r-s | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
- GFP-DT(positive control)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
Transformation
- LacP-torA-GFP-DT(Backbone pSB3C5)
- J23107-TatABCD
PCR
kil, torA-GFP
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→25cycles
August 22
Restrictive Digestion
| pSB3C5 | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 13.3 | 0.5 | 0.5 | 3 | 0.5 | 12.2 | 30 |
Gel extraction
pSB3C5(EcoR1, Pst1)
27.2μg/mL
Transformation(the second time)
- LacP-torA-GFP-DT(Backbone pSB3C5)
- J23107-TatABCD
Electrophoresis
See "August 21 -Colony PCR-"
1. 1kb ladder
2. pspA(Primer pspA f&r)
3. negative control for 2
4. GFP-DT
5. pspA(Primer pspA f-p&r-s)
6. pspA(Primer pspA f-p&r-s)
7. negative control for 5&6
PCR
LacP-GFP-DT(a kit of biological parts)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
Colony PCR
pspA
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer pspA f | Primer pspA r | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | Primer pspA f-p | Primer pspA r-s | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→30cycles
Electrophoresis
1. 1kb ladder
2. pspA(Primer pspA f&r)
3. pspA(Primer pspA f-p&r-s)
4. negative control for pspA(Primer pspA f&r)
5. negative control for pspA(Primer pspA f-p&r-s)
Transformation
Kil
August 23
Colony PCR
J23107-TatABCD
VF and VR were used for amplifying approximately 2800bp
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32.5 | 0.5 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 90sec
→25cycles
August 24
Purification of PCR products
pspA(Primer pspA f-p&r-s)
84.1μg/mL
→See "August 22 -Colony PCR-"
Restrictive Digestion
| pspA | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
| pspA | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1h
Gel extraction
- pspA(EcoR1, Pst1) 30.7μg/mL
- pspA(Xba1, Pst1) 44.0μg/mL
Restrictive Digestion
| pSB1C3(82.9μg/mL) | EcoR1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
→Gel extraction
Electrophoresis
1. 1kb ladder
2. J23107-TatABCD →See "August 23 -PCR-"
3. negative control for J23107-TatABCD
4. pSB1C3(EcoR1, Pst1)
Ligation
| pSB3C5(EcoR1, Pst1) | pspA(EcoR1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
16℃, overnight
August 25
August 26
August 27
Ligation
| pSB1C3(EcoR1, Pst1) | pspA(EcoR1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
16℃, overnight
Colony PCR
Kil (1-7)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→30cycles
Purification of PCR products
torA-GFP →See "August 21 -PCR-"
Restrictive Digestion
| LacP(Backbone:pSB3C5) | Spe1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1h
→Gel extraction
| torA-GFP | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1h
→Purification
Purification of PCR products
J23107-TatABCD
42.0μg/mL
→See "August 23 -PCR-"
Restrictive Digestion
| J23107-TatABCD | Spe1 | EcoR1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
August 28
Colony PCR
Kil (1-16)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
Transformation
- pspA-pSB1C3
- pspA-pSB3C5
Liquid culture
- Kil-3 →Miniprep 100.4μg/mL
- Kil-6
- Kil-7 →Miniprep 77.7μg/mL
- LacP-torA-GFP-1 →Miniprep 88.3μg/mL
- LacP-torA-GFP-2 →Miniprep 85.0μg/mL
- LacP-torA-GFP-3 →Miniprep +++
August 29
Kil assay
After culturing for 18hr at 37℃, we eliminated the supernatant using a centrifuge, and diluted it until OD600=0.1. Then we dispensed it. The dispense volume was 3mL. We added 0/0.001/0.01/0.1/1mM IPTG to each. While culturing again at 37℃, we measured OD600.
| IPTG | 0 | 0.001mM | 0.01mM | 0.1mM | 1mM |
|---|---|---|---|---|---|
| 0.5h | 0.202 | 0.207 | 0.201 | 0.207 | 0.200 |
| 1h | 0.406 | 0.428 | 0.424 | 0.402 | 0.421 |
| 1.5h | 0.796 | 0.813 | 0.779 | 0.751 | 0.802 |
| 2h | 1.107 | 1.129 | 1.141 | 1.092 | 1.124 |
| 2.5h | 1.546 | 1.565 | 1.541 | 1.532 | 1.578 |
| 3h | 1.912 | 1.933 | 1.890 | 1.883 | 1.940 |
| 3.5h | 2.300 | 2.295 | 2.238 | 2.247 | 2.259 |
| 4h | 2.546 | 2.545 | 2.528 | 2.490 | 2.520 |
| 4.5h | 2.688 | 2.663 | 2.603 | 2.633 | 2.666 |
| 5h | 2.699 | 2.826 | 2.787 | 2.593 | 2.673 |
| 5.5h | 2.863 | 2.742 | 2.754 | 2.741 | 2.756 |
| 6h | 2.831 | 2.876 | 2.758 | 2.759 | 2.728 |
| 20h | 2.671 | 2.706 | 2.680 | 2.606 | 2.619 |
Liquid culture
- pspA-pSB1C3(1-3)
- pspA-pSB3C5(1-3)
Restrictive Digestion
| LacP-Kil-DT(134.7ng/μL) | EcoR1 | Spe1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 15 | 30 |
37℃, overnight
Colony PCR
- LacP-torA-GFP-DT (1-5)
- pspA-pSB1C3 (1-5)
- pspA-pSB3C5 (1-6)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 40sec
→30cycles
Gel extraction
J23107-TatABCD(EcoR1, Pst1) -5.9μg/mL
August 30
Ligation
| J23107-TatABCD(EcoR1, Spe1)22.2μg/mL | DT(EcoR1, Xba1)37.5μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 1 | 9 | 20 |
at 16℃ for 1h
Transformation
J23107-TatABCD-DT (Amp resistance)
Electrophoresis
1. LacP-torA-GFP-DT-1
2. LacP-torA-GFP-DT-2
3. LacP-torA-GFP-DT-3
4. LacP-torA-GFP-DT-4
5. LacP-torA-GFP-DT-5
6. pspA-pSB1C3-1
7. pspA-pSB1C3-2
8. pspA-pSB1C3-3
9. pspA-pSB1C3-4
10.pspA-pSB1C3-5
11.pspA-pSB3C5-1
12.pspA-pSB3C5-2
13.pspA-pSB3C5-3
14.pspA-pSB3C5-4
15.pspA-pSB3C5-5
16.pspA-pSB3C5-6
Miniprep
- pspA-pSB3C5-1 176μg/mL
- pspA-pSB3C5-2 214μg/mL
- pspA-pSB3C5-3 461μg/mL
- pspA-pSB1C3-2 79μg/mL
Colony PCR
LacP-torA-GFP-DT (6-13)
| 2×Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
56℃, 30sec
68℃, 1min
→30cycles
Restrictive Digestion
| pspA-pSB1C3 | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 5 | 0.5 | 28.5 | 50 |
at 37℃ for 1h
Electrophoresis
1. 1kb ladder
3. pspA-pSB1C3(EcoR1, Spe1)
August 31
Gel extraction
pspA-pSB1C3(EcoR1, Spe1) 8.0μg/mL
Purification
LacP-GFP-DT -0.7μg/mL
Ligation
| pspA(EcoR1, Spe1) | DT(EcoR1, Xba1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 25 | 1 | 13 | 39 |
at 16℃ for 1h
Transformation
pspA-DT (Amp resistance)
September 2
Colony PCR
J23107-TatABCD-DT (1-8)
| 2×Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 3min
→25cycles
pspA-DT (2-8)
| 2×Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Extension, 10sec
→25cycles
Restrictive Digestion
| LacP-torA-GFP-DT | EcoR1 | Spe1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 2 | 6 | 20 |
at 4℃ for overnight
Electrophoresis
1. 1kb ladder
3. J23107-TatABCD-DT-1
4. J23107-TatABCD-DT-3
5. J23107-TatABCD-DT-4
6. J23107-TatABCD-DT-5
7. J23107-TatABCD-DT-6
8. J23107-TatABCD-DT-7
9. J23107-TatABCD-DT-8
1. 1kb ladder
3. J23107-TatABCD-DT-1
4. J23107-TatABCD-DT-3
5. J23107-TatABCD-DT-4
6. J23107-TatABCD-DT-5
7. J23107-TatABCD-DT-6
8. J23107-TatABCD-DT-7
9. J23107-TatABCD-DT-8
11. LacP-pSB3C5(Spe1, Pst1)
1. 1kb ladder
3. pspA-DT-2
4. pspA-DT-3
5. pspA-DT-4
6. pspA-DT-5
7. pspA-DT-6
8. pspA-DT-7
9. pspA-DT-8
September 3
PCR
TatABCD-DT
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 32 | 1 | 50 |
Gel extraction
LacP-torA-GFP-DT →See "September 5 -Gel extraction-"
Restrictive Digestion
| torA-GFP-DT | Xba1 | Pst1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 20 | 2 | 2 | 4 | 12 | 40 |
Purification
J23107-TatABCD 29.6μg/mL 1.56(260/280) 1.60(260/230)
Electrophoresis
1. 1kb ladder
2. J23107-TatABCD
Liquid culture
- LacP-kil-DT(1)
- pspA-DT(4, 7)
PCR
J23107-TatABCD
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | DNA | Total |
|---|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 29 | 4 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 10sec
→30cycles
Gel extraction
LacP-torA-GFP-DT 31μg/mL 1.28(260/280) 0.49(260/230)
LacP-torA-GFP-DT 8μg/mL 1.32(260/280) 0.83(260/230)
Transformation
J23107-TatABCD (Amp resistance)
J23113-kil-DT (Amp resistance)
J23100-kil-DT (CP resistance)
J23101-kil-DT (CP resistance)
J23106-kil-DT (CP resistance)
J23115-kil-DT (CP resistance)→We didn't transform because of lack of culture medium.
J23105-kil-DT (CP resistance)→We didn't transform because of lack of culture medium.
Electrophoresis
1. 1kb ladder
2-7. LacP-torA-GFP-DT
Colony PCR
LacP-torA-GFP-DT (14-21) J23113-kil-DT (Amp resistance, 1-4) J23113-kil-DT (CP resistance, 1-4)
| 2×Quick Taq | primer-f | primer-r | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 1min
→25cycles
September 4
Liquid culture
- LacP-kil-DT(4, 5)(Amp resistance)
- pspA-DT(3, 6, 7)(Amp resistance)
PCR
We did PCR 10 more cycles to amplify products of PCR that we had done yesterday because we could not amplify J23107-TatABCD on September 3.
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 1.5min
→30cycles
Electrophoresis

1-8. LacP-torA-GFP-DT

1-4. J23113-kil-DT (Amp resistance)
5-8. J23113-kil-DT (CP resistance)
Liquid culture
- LacP-torA-GFP-DT
- J23113-kil-DT
- LacP-kil-DT(with IPTG)
- LacP-kil-DT(without IPTG)
After 20 hour, we quantified OD600.
| plasmid | OD600 |
|---|---|
| LacP-kil-DT(with IPTG) | 2.116 |
| LacP-kil-DT(without IPTG) | 2.320 |
We cultured CP tolerance plasmid and give an antibiotic(Amp or Kan or none) after 2 hours. We quantify OD600.
| time | 1 | 2 | 3 |
|---|---|---|---|
| 2hours | 0.131 | 0.100 | 0.606 |
| add an antibiotic | Amp | Kan | none |
| 7hours | 0.491 | 0.988 | 2.634 |
| 19hours | 2.253 | 0.815 | 2.742 |
September 5
Colony PCR
J23107-TatABCD (1-7)
| buffer | 2mM dNTPs | 25mM MgSO4 | primer-f | primer-r | KOD Plus Neo | DW | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 1.25min
→30cycles
Gel extraction
by Kato
See "September 3 -Gel extraction-"
LacP-torA-GFP-DT 15.9μg/mL 1.29(260/280) 0.38(260/230)
LacP-torA-GFP-DT 17.2μg/mL 1.23(260/280) 0.48(260/230)
Miniprep
- J23113-kil-DT 85.7μg/mL 1.76(260/280) 1.97(260/230)
- LacP-torA-GFP-DT 126.6μg/mL 1.81(260/280) 2.09(260/230)
- LacP-kil-DT-4 62.5μg/mL 1.79(260/280) 1.99(260/230)
- LacP-kil-DT-5 56.0μg/mL 1.79(260/280) 2.01(260/230)
- pspA-DT-3 322μg/mL 1.45(260/280) 1.42(260/230)
- pspA-DT-6 147.6μg/mL 1.25(260/280) 1.37(260/230)
- pspA-DT-7 145.5μg/mL 1.24(260/280) 1.27(260/230)
Restrictive Digestion
| pspA-DT-6 | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
at 37℃ for 1h
Electrophoresis
J23107-TatABCD (1-7)
Colony PCR
J23107-TatABCD (8-23)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 1.5min
→30cycles
Gel extraction
pspA-DT 10.2μg/mL 1.42(260/280) 0.76(260/230)
Ligation
| pspA-DT(Xba1, Pst1) | LacP(pSB3C5)(Spe1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 5 | 1 | 3 | 9 |
at 16℃
Electrophoresis

LacP-torA-GFP-DT(8-15)
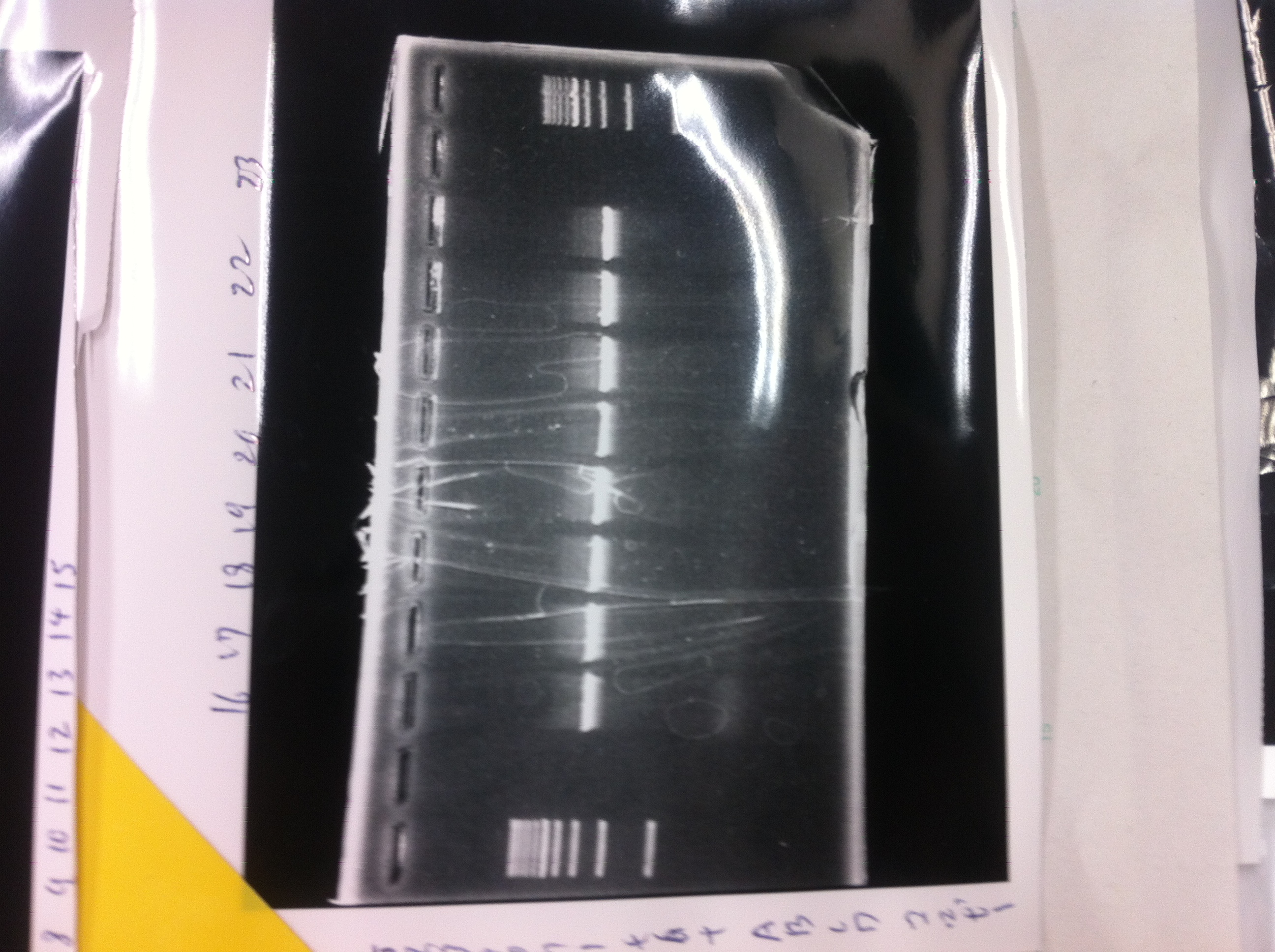
LacP-torA-GFP-DT(9-23)
Ligation
| LacP-torA-GFP-DT(EcoR1, Spe1) | DT 17.2μg/mL(Xba1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 15 | 25 | 20 | 60 |
Liquid culture
- J23107-TatABCD-8
- J23107-TatABCD-20
- LacP-torA-GFP-DT-21
September 6
Miniprep
- J23107-TatABCD-8 82.4μg/mL 2.06(260/280) 2.95(260/230)
- J23107-TatABCD-20 102.9μg/mL 2.04(260/280) 2.94(260/230)
Restrictive Digestion
| J23107-TatABCD 102.9μg/mL | Spe1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
at 37℃ for 1h
Transformation
LacP-pspA-DT(pSB3C5)
| DNA | Competent cell | Total |
|---|---|---|
| 2 | 20 | 22 |
Gel extraction
J23107-TatABCD(Spe1, Pst1)
→We failed gel extraction because we mistook steps of experiment.
Electrophoresis
LacP-torA-GFP-DT-pspA-DT
Restrictive Digestion
by Terasaka
| J23107-TatABCD | Spe1 | Pst1 | BufferH | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
Gel extraction
J23107-TatABCD(Spe1, Pst1)
→See "September 7 -Gel extraction-"
September 7
Liquid culture
- LacP-torA-GFP-DT-1 (CP resistance)
- LacP-torA-GFP-DT-21 (CP resistance)
preculture for 1 hour
Colony PCR
LacP-torA-GFP-DT (1-8) LacP-pspA-DT (1-8)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 1.5min
→30cycles
Restrictive Digestion
by Terasaka
| J23107-TatABCD | EcoR1 | Spe1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
| GFP-DT | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 3 | 0.5 | 10.5 | 30 |
Electrophoresis
① -⑧. LacP-pspA-DT
1-8. LacP-torA-GFP-DT
Gel extraction
- J23107-TatABCD(EcoR1, Spe1)
- J23107-TatABCD(Pst1, Spe1) ①57.6μg/mL ②23.4μg/mL
- GFP-DT(Xba1, Pst1)
Restrictive Digestion
| pspA-DT(145.5μg/mL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 20 | 0.5 | 0.5 | 3 | 0.5 | 5.5 | 30 |
37℃, overnight
Liquid culture
- LacP-torA-GFP-DT-7 (LB 2mL)
+IPTG 0mM, 0.05mM, 0.1mM, 0.15mM, 0.2mM, 0.3mM
- LacP-torA-GFP-DT-7 (LB 5mL)
September 8
Check the effect of Amp
We used 5mL of the liquid culture medium with LacP-torA-GFP-DT-7.
→See "September 7 -Liquid culture-"
We diluted it with LB until OD600=0.444, and dispensed it.
The dispense volume was 1.3mL.
We added 0/13/130μL of Amp(50mg/mL) to it.
5.5h after incubating at 37℃
| Amp | 0μL | 13μL | 130μL |
|---|---|---|---|
| OD600 | 2.457 | 2.696 | 0.310 |
Observation through a confocal microscope
We used the liquid culture media with LacP-torA-GFP-DT + 13μL of Amp(50mg/mL) + IPTG 0/0.1/0.15mM.
Their OD600 was 0.47.
3h after incubating at 37℃, we eliminated each supernatant using a centrifuge, and diluted them with LB to which Amp was added.
2.5h after incubating at 37℃, we observed them through a confocal microscope.
We failed to observe it.
Gel extraction
pspA-DT -5.5μg/mL
Restrictive Digestion
| pspA-DT-3(322μg/mL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 3 | 12 | 30 |
at 37℃ for 1h
September 9
Restrictive Digestion
| J23107-TatABCD(102.9μg/mL) | EcoR1 | Spe1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 15 | 30 |
Electrophoresis
1. J23107-TatABCD(EcoR1, Spe1)
2. J23107-TatABCD(before restriction)
3. pspA-DT(Xba1, Pst1)
4. pspA-DT(before restriction)
Gel extraction
- J23107-TatABCD(EcoR1, Spe1) 26μg/mL
- pspA-DT(Xba1, Pst1) 199μg/mL
Liquid culture
- J23100/J23101/J23106-1
- pspA-DT-5 ×2
- LacP-torA-GFP-DT-21
- GFP generator-2
- LacP-pspA-DT-6
September 10
Miniprep
- J23100-1 105.5μg/mL
- J23101-1 76.0μg/mL
- J23106-1 90.9μg/mL
- pspA-DT-5 ①104.1μg/mL ②80.0μg/mL
- LacP-torA-GFP-DT-21 79.6μg/mL
- GFP generator-2 84.1μg/mL
- LacP-pspA-DT-6 99.8μg/mL
Ligation
- J23107-TatABCD-DT
| J23107-TatABCD(EcoR1, Spe1) | DT(EcoR1, Xba1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 5 | 7.5 | 22.5 |
- Negative control for J23107-TatABCD-DT
| DT(EcoR1, Xba1) | Ligation High Ver.2 | Total |
|---|---|---|
| 5 | 5 | 10 |
- J23107-TatABCD-pspA-DT
| J23107-TatABCD(Spe1, Pst1) | pspA-DT(Xba1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 3 | 6 | 9 | 18 |
- Negative control for J23107-TatABCD-pspA-DT
| J23107-TatABCD(Spe1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|
| 3 | 3 | 6 |
Restrictive Digestion
| DNA | Spe1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
DNA=J23100(105.5μg/mL), J23101(76.0μg/mL), J23106(90.9μg/mL)
at 37℃
Gel extraction
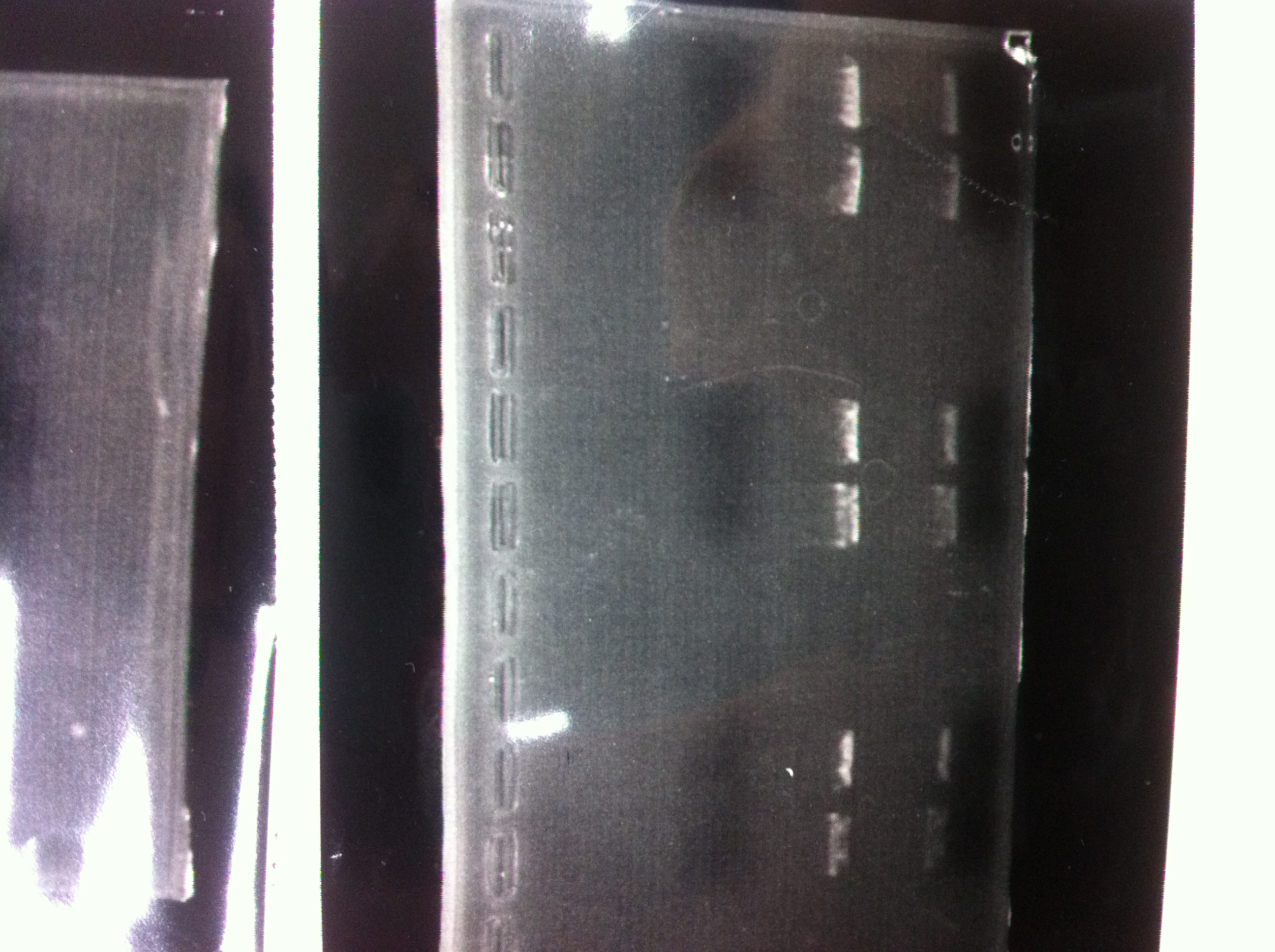
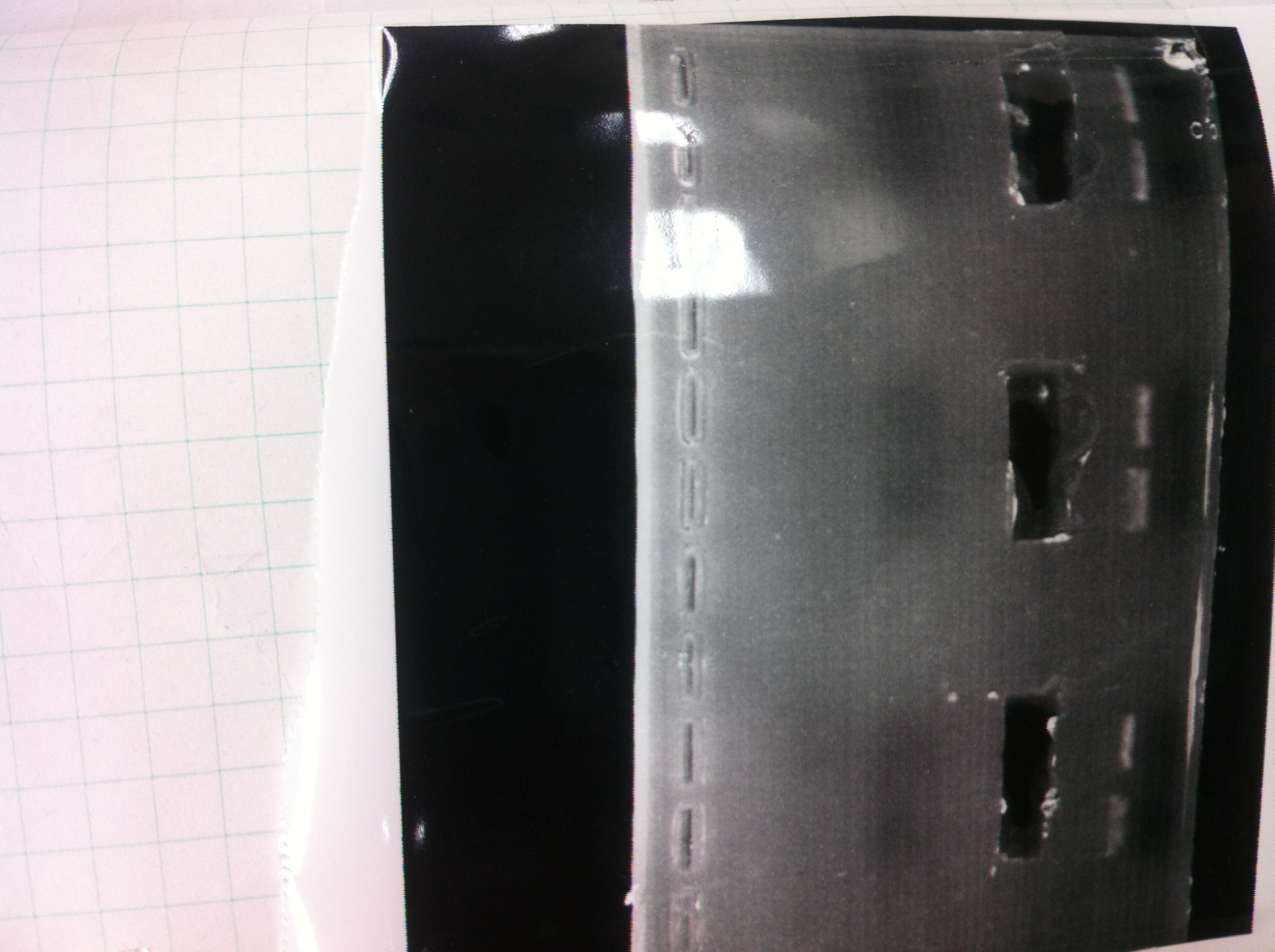
J23100 27.1μg/mL
J23101 50.2μg/mL
J23106 16.1μg/mL
Ligation
- J23100/J23101/J23106-kil-DT
| J23100/J23101/J23106(Spe1, Pst1) | kil-DT(Xba1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
- Negative control for J23100/J23101/J23106-kil-DT
| J23100/J23101/J23106(Spe1, Pst1) | MilliQ | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
Transformation
J23100/J23101/J23106-kil-DT
September 11
Colony PCR
- J23107-TatABCD-DT (1-8)
- J23107-TatABCD-pspA-DT (1-8)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 3.5min
→30cycles
Electrophoresis
- J23107-TatABCD-DT (1-8)
- J23107-TatABCD-pspA-DT (1-8)
Restrictive Digestion
| GFP generator(84.1μg/mL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 3 | 7 | 30 |
| LacP-torA-GFP-DT(85.0μg/mL) | Spe1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
| LacP-pspA-DT(99.8μg/mL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 3 | 7 | 30 |
Liquid culture
pSB1C3(self-ligation) ×5 in LB 3mLCP 3μL
When OD600=approximately 0.6, we added 0/30/60/150/300μL of Amp(50mg/mL) to each liquid culture medium.
1h and 5h after incubating at 37℃, we measured OD600.
| 1h | 5h | |
|---|---|---|
| 0 | -0.020 | 0.403 |
| 30 | -0.046 | 0.544 |
| 60 | 0.263 | 0.007 |
| 150 | 0.061 | 0.839 |
| 300 | 0.093 | 1.441 |
Ligation
| pSB3C5(EcoR1, Pst1) | pspA-DT(Xba1, Pst1) | J23107-TatABCD(EcoR1, Spe1) | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 1 | 3 | 5 | 4.5 | 13.5 |
4℃, overnight
Colony PCR
- J23100-kil-DT (1-6)
- J23101-kil-DT (1-7)
- J23106-kil-DT (1-2)
| 2×Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 23 | 1 | 1 | 25 | 50 |
Gel extraction
- GFP generator (Xba1, Pst1) 14.1μg/mL
- LacP-torA-GFP-DT (Spe1, Pst1) 42.9μg/mL 1.13(260/280) 1.19(260/230)
- LacP-pspA-DT (Xba1, Pst1) 20.0μg/mL 1.01(260/280) 1.30(260/230)
Electrophoresis
- J23107-TatABCD-DT (1-8)
- J23106-kil-DT (1, 2)
- J23100-kil-DT (1-4)
Liquid culture
- LacP-torA-GFP-DT
- pSB1C3
Colony PCR
We did PCR 10 more cycle to amplify products of PCR on 9/11 of J23107-TatABCD-pspA-DT and J23107-TatABCD-DT.
September 12
Colony PCR
J23107-TatABCD-pspA-DT (9-15)
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 4min
→26cycles
Check the effect of Amp
We cultured pSB1C3 for overnight.
→See "September 7 -Liquid culture-"
We diluted it with LB, and dispensed it.
We added Amp(50mg/mL) to it until density of Amp was 1/10, 1/30, 1/50 .
| Amp | start | 5 hours |
|---|---|---|
| 1/10 | 0.557 | 0.077 |
| 1/30 | 0.628 | 0.166 |
| 1/50 | 0.472 | 0.055 |
| 0 | 0.594 | 2.585 |
Electrophoresis
- J23107-TatABCD-pspA-DT
- J23101-kil-DT (1-7)
Restrictive Digestion
| pspA(pSB3C5) (106μg/mL) | EcoR1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
| pspA-DT-3 | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 3 | 7 | 30 |
| J23107-TatABCD (102.4μg/mL) | EcoR1 | Spe1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
Electrophoresis
- pspA(pSB3C5)(EcoR1, Pst1)
- pspA-DT(Xba1, Pst1)
- J23107-TatABCD(EcoR1, Spe1)
- pSB1C3(EcoR1, Spe1)
Transformation
J23107-TatABCD-pspA-DT(pSB3C5)
| DNA | Competent cell | Total |
|---|---|---|
| 2 | 20 | 22 |
on ice for 30 minites heatshock at 42℃ for 1 hour on ice 2 minites then add SOC medium 200μl preculture at 37℃ for 1 hour incubate CP culture plate
Gel extraction
- pspA(pSB3C5)(EcoR1, Pst1) 26.8μg/mL 1.27(260/280) 0.32(260/230)
- pspA-DT(Xba1, Pst1) 26.1μg/mL 1.13(260/280) 0.55(260/230)
- J23107-TatABCD(EcoR1, Spe1) 1.5μg/mL 1.99(260/280) 0.06(260/230)
Ligation
| pSB3C5(EcoR1, Pst1) 26.8μg/mL | pspA-DT(Xba1, Pst1) 26.1μg/mL | J23107-TatABCD(EcoR1, Spe1) 1.5μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 1 | 3 | 6 | 5 | 15 |
| pSB3C5(EcoR1, Pst1) 26.8μg/mL | MilliQ | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 9 | 5 | 15 |
| LacP(pSB3C5)(Spe1, Pst1) 10μg/mL | GFP-DT(Xba1, Pst1) 7.0μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
| LacP(pSB3C5)(Spe1, Pst1) 10μg/mL | MilliQ | Ligation High Ver.2 | Total |
|---|---|---|---|
| 1 | 5 | 3 | 9 |
4℃, overnight
Observation through a confocal microscope
left:E.coli sample stained with Hoechstunder and observed with 352nm wavelength
right:E.coli having Lacp-torA-GFP-DT and observed with 489nm wavelength
September 13
Miniprep
- J23107-TatABCD-pspA-DT(pSB3C5)-1 130μg/mL 1.37(260/280) 0.79(260/230)
Restrictive Digestion
| J23107-TatABCD-pspA-DT(pSB3C5) (130μg/mL) | Spe1 | Pst1 | BufferH | MilliQ | Total
|
|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 15 | 30 |
Transformation
- LacP-GFP generater
- LacP
- J23107-TatABCD-pspA-DT(pSB3C5)
- J23107-TatABCD
Colony PCR
J23107-TatABCD-pspA-DT
→See "September 14 -Colony PCR-"
September 14
Colony PCR
See "September 13 -Colony PCR-"
J23107-TatABCD-pspA-DT
Colony PCR
- J23107-TatABCD-pspA-DT (1-6)
| 2×Quick Taq | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 23 | 1 | 1 | 25 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 4min
→30cycles
September 15
Liquid culture
- J23107-TatABCD-pspA-DT-1
- J23107-TatABCD-pspA-DT-2
- J23107-TatABCD-pspA-DT-3
Miniprep
- J23107-TatABCD-pspA-DT-1 22.9μg/mL
- J23107-TatABCD-pspA-DT-2 51.8μg/mL 1.57(260/280) 1.01(260/230)
- J23107-TatABCD-pspA-DT-3 28.5μg/mL 1.57(260/280) 0.94(260/230)
Restrictive Digestion
| J23107-TatABCD-pspA-DT (51.8μg/mL) | EcoR1 | Spe1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 3 | 11 | 30 |
| LacP-torA-GFP-DT (79.6μg/mL) | EcoR1 | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 0.5 | 0.5 | 3 | 0.5 | 15.5 | 30 |
| LacP-kil-DT (56.0μg/mL) | EcoR1 | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 3 | 0.5 | 10.5 | 30 |
| J23107-TatABCD-pspA-DT (51.8μg/mL) | EcoR1 | Pst1 | BufferM | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 0.5 | 0.5 | 3 | 11 | 30 |
37℃, 2 hours
Electrophoresis
①. 1kb ladder
②. J23107-TatABCD-pspA-DT
③. J23107-TatABCD-pspA-DT(EcoR1, Spe1)
④. J23107-TatABCD-pspA-DT(EcoR1, Pst1)
⑤. LacP-kil-DT
⑥. LacP-kil-DT(EcoR1, Xba1)
⑦. LacP-torA-GFP-DT
⑧. LacP-torA-GFP-DT(EcoR1, Xba1)
Gel extraction


①. 1kb ladder
②. J23107-TatABCD-pspA-DT(EcoR1, Spe1)
③. J23107-TatABCD-pspA-DT(EcoR1, Spe1)
④. J23107-TatABCD-pspA-DT(EcoR1, Pst1)
⑤. J23107-TatABCD-pspA-DT(EcoR1, Pst1)
⑥. 1kb ladder
- J23107-TatABCD-pspA-DT(EcoR1, Spe1) 14.2μg/mL 1.28(260/280) 0.54(260/230)
- J23107-TatABCD-pspA-DT(EcoR1, Pst1) 7.3μg/mL 1.55(260/280) 0.38(260/230)
Purification
- LacP-torA-GFP-DT(EcoR1, Xba1) 7.7μg/mL 1.41(260/280) 0.99(260/230)
- LacP-kil-DT(EcoR1, Xba1) →failed
Ligation
| LacP-torA-GFP-DT(EcoR1, Xba1) | J23107-TatABCD-pspA-DT(EcoR1, Spe1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 10 | 7 | 21 |
| pSB1C3(EcoR1, Pst1) | J23107-TatABCD-pspA-DT(EcoR1, Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 4 | 10 | 7 | 21 |
at 16℃ for overnight
September 16
Transformation
LacP-torA-GFP-DT-J23107-TatABCD-pspA-DT
J23107-TatABCD-pspA-DT(pSB1C3)
September 17
Colony PCR
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (1-6)
J23107-TatABCD-pspA-DT(pSB1C3) (1-6)
J23107-kil-DT (6-10)
Restrictive Digestion
| LacP-torA-GFP-DT (126μg/mL) | EcoR1 | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 3 | 12 | 30 |
| LacP-torA-GFP-DT (126μg/mL) | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 3 | 1 | 3 | 3 | 20 | 30 |
Electrophoresis
A1-A6. J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (1-6)
B1-B6. J23107-TatABCD-pspA-DT(pSB1C3) (1-6)
Liquid culture
- J23107-TatABCD-pspA-DT(pSB1C3)
- J23107-TatABCD-pspA-DT(pSB3C5)
- LacP-torA-GFP-DT(pSB3C5)
Electrophoresis
lane1. 1kb ladder
lane2. J23101-kil-DT-6
lane3. J23101-kil-DT-7
lane4. J23101-kil-DT-8
lane5. J23101-kil-DT-10
lane6. LacP-torA-GFP-DT
lane7. LacP-torA-GFP-DT(EcoR1, Xba1)
lane8. LacP-torA-GFP-DT(Xba1)
Miniprep
- J23106-kil-DT 11.7μg/mL 1.93(260/280) 1.46(260/230)
September 18
Gel extraction
- LacP-torA-GFP-DT(EcoR1, Xba1) 36.9μg/mL 1.37(260/280) 0.82(260/230)
Ligation
| J23107-TatABCD-pspA-DT(EcoR1, Spe1) 14.2μg/mL | LacP-torA-GFP-DT(EcoR1, Xba1) 36.9μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|
| 8 | 1.5 | 9.5 | 19 |
| LacP-torA-GFP-DT(EcoR1, Xba1) 36.9μg/mL | Ligation High Ver.2 | Total |
|---|---|---|
| 1.5 | 1.5 | 3 |
at 4℃ for 1 hour
Transformation
- J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT
- LacP-torA-GFP-DT(EcoR1, Xba1)
Observation through a confocal microscope
We made sample for observation through a confocal microscope.
We incubated sample with 0mM/0.5mM IPTG for 1.5 hours.
Then, we incubated onto medium without IPTG for 2.5 hours.
| IPTG(mM) | medium |
|---|---|
| 0 | LB |
| 0.1 | LB |
| 0.1 | LB with 0.1mM IPTG (positive control) |
| 0 | LB with 2 percent glucose |
| 0.1 | LB with 2 percent glucose |
| 0 | LB with 5 percent glucose |
| 0.1 | LB with 5 percent glucose |
| 0 | LB with 2 percent glucose (make medium new after incubate for 1.5 hours) |
| 0 | LB with 2 percent glucose (make medium new after incubate for 1.5 hours) |
Restrictive Digestion
| pSB4K5 | EcoR1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 15 | 30 |
| pSB4K5 | EcoR1 | BufferH | MilliQ | Total |
|---|---|---|---|---|
| 3 | 0.5 | 1 | 5.5 | 30 |
| pSB4K5 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|
| 3 | 0.5 | 1 | 5.5 | 10 |
| LacP-torA-GFP-DT | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 3 | 12 | 30 |
| LacP-torA-GFP-DT | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 2 | 1 | 1 | 1 | 5 | 10 |
| LacP-torA-GFP-DT | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|
| 2 | 1 | 1 | 6 | 10 |
37℃, 2 hours
Gel extraction
- LacP-torA-GFP-DT(Xba1, Pst1) 9.7μg/mL 1.34(260/280) 0.62(260/230)
- pSB4K5(EcoR1, Pst1) 9.6μg/mL 1.28(260/280) 0.32(260/230)
Miniprep
- J23107-TatABCD-psp-DT(pSB1C3) 78μg/mL
- J23107-TatABCD-psp-DT(pSB1C3) 48μg/mL
- LacP-torA-GFP-DT 29μg/mL
- J23101-kil-DT 25μg/mL
Restrictive Digestion
| J23101-Kil-DT 25µg/ml | BufferM | Xba1 | Pst1 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 40 | 6 | 1 | 1 | 6 | 6 | 60 |
| J23101-Kil-DT 25µg/ml | BufferM | Xba1 | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 3 | 1 | 0.5 | 1 | 4.5 | 10 |
| J23101-Kil-DT 25µg/ml | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 3 | 1 | 0.5 | 5.5 | 60 |
at 37℃ for 2 hours
Electrophoresis
①. pSB4K5
②. pSB4K5(Pst1)
③. pSB4K5(EcoR1)
④. pSB4K5(EcoR1, Pst1)
⑤. LacP-torA-GFP-DT
⑥. LacP-torA-GFP-DT(Xba1)
⑦. LacP-torA-GFP-DT(Pst1)
⑧. LacP-torA-GFP-DT(Xba1, Pst1)
①. 1kb ladder
②. J23101-Kil-DT(Xba1, Pst1)
③. J23101-Kil-DT(Xba1, Pst1)
④. J23101-Kil-DT(Xba1, Pst1)
⑤. J23101-Kil-DT(Xba1, Pst1)
⑥. 1kb ladder
Restrictive Digestion
| J23101-Kil-DT | BufferM | Xba1 | Pst1 | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 3 | 1 | 1 | 3 | 7 | 30 |
| J23101-Kil-DT | BufferM | Xba1 | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 3 | 1 | 0.5 | 1 | 4.5 | 10 |
| J23101-Kil-DT | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 3 | 1 | 0.5 | 5.5 | 10 |
37℃
September 19
Gel extraction
lane1. 1kb ladder
lane2. pSB4K5(EcoR1, Pst1)
lane3. pSB4K5(EcoR1, Pst1)
lane4. empty
lane5. LacP-torA-GFP-DT(Xba1, Pst1)
lane6. LacP-torA-GFP-DT(Xba1, Pst1)
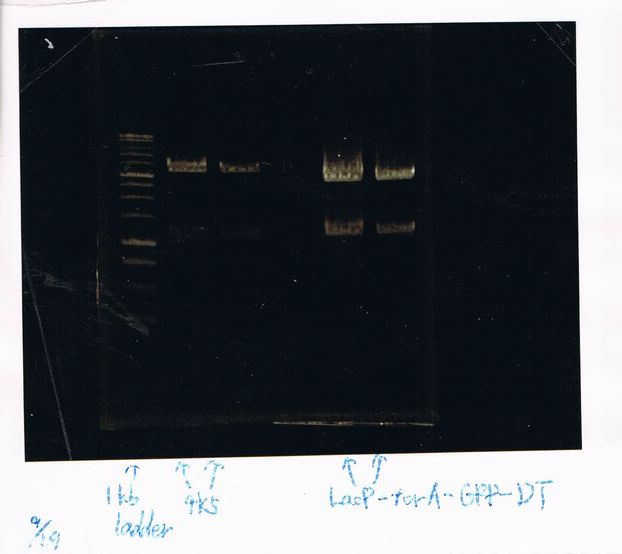
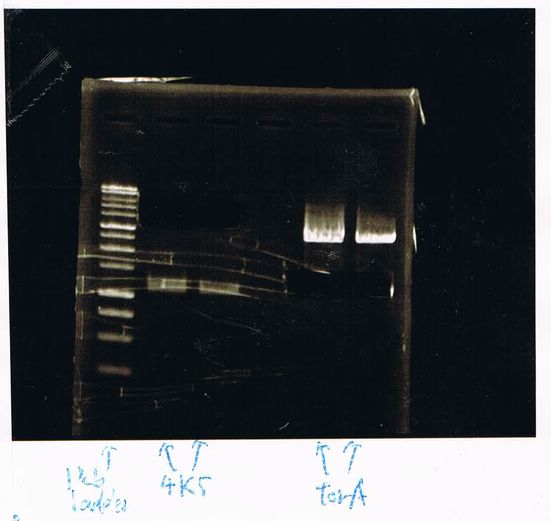
Ligation
| LacP-torA-GFP-DT(Xba1,Pst1) 9.7μg/mL | pSB4K5(EcoR1,Pst1) 9.6μg/mL | J23107-TatABCD-PsPA-DT(EcoR1,Spe1) 14.2μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 7 | 2 | 7 | 8 | 24μL |
| pSB4K5(EcoR1,Pst1) 9.6μg/mL | Ligation High Ver.2 | MilliQ | Total |
|---|---|---|---|
| 2 | 1 | 21 | 24μL |
incubate at 37℃ for 2h
Electrophoresis
J23101-kil-DT (Xba1,Pst1), (Xba1), (Pst1)
Lane1: 1kb ladder
Lane2: empty
Lane3: plasmid
Lane4: (Xba1)
Lane5: (Pst1)
Lane6: (Xba1, Pst1)
Liquid Culture
We did liquid culture J23106-kil-DT(9/10, Amp)'s 6,7 at LB mediumAmp.
Transformation
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT
pSB4K5 negative control
Liquid culture
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:1~5
Colony PCR
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT
| VF2 | VR | Quick tag | MilliQ | Total |
|---|---|---|---|---|
| 1 | 1 | 25 | 23 | 50 |
Predenature 94℃ 2min
Denature 94℃ 30sec
Annealing 55℃ 30sec
Extension 68℃ 5min
→30cycles
Restrictive Digestion
| LacP-kil-DT(4) 62.5μg/mL | Xba1 | Pst1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|---|
| 10 | 1 | 1 | 3 | 12 | 3 | 30 |
| LacP-kil-DT(4) | EcoR1 | Pst1 | MilliQ | BufferH | Total |
|---|---|---|---|---|---|
| 10 | 1 | 1 | 15 | 3 | 30 |
| LacP-kil-DT | Xba1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|
| 2 | 0.5 | 1 | 5.5 | 1 | 10 |
| LacP-kil-DT | EcoR1 | MilliQ | BufferH | Total |
|---|---|---|---|---|
| 2 | 0.5 | 6.5 | 1 | 10 |
| LacP-kil-DT | Pst1 | MilliQ | BufferH | Total |
|---|---|---|---|---|
| 2 | 0.5 | 6.5 | 1 | 10 |
at 37℃, 2 hours
Electrophoresis
LacP-kil-DT
Lane1: 1kb ladder
Lane2: empty
Lane3: plasmid
Lane4: (EcoR1)
Lane5: (Xba1)
Lane6: (Pst1)
Lane7: (EcoR1, Pst1)
Lane8: (Xba1, Pst1)
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(1-5)
Lane1: ladder
Lane2: 1
Lane3: 2
Lane4: 3
Lane5: 4
Lane6: 5
Ligation
| LacP-torA-GFP-DT(Xba1,Pst1) 9.7μg/mL | pSB4K5(EcoR1,Pst1) 9.6μg/mL | J23107-TatABCD-PsPA-DT(EcoR1,Spe1) 14.2μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 7 | 4 | 7 | 9 | 27μL |
(Negative control)
| pSB4K5(EcoR1,Pst1) 9.6μg/mL | Ligation High Ver.2 | MilliQ | Total |
|---|---|---|---|
| 4 | 9 | 14 | 27μL |
Restrictive Digestion
| torA-R9 101.8μg/mL | bufferH | MilliQ | EcoR1 | Pst1 | Total |
|---|---|---|---|---|---|
| 10 | 3 | 15 | 1 | 1 | 30 |
| torA-R9 101.8μg/mL | bufferH | MilliQ | EcoR1 | Pst1 | Total |
|---|---|---|---|---|---|
| 3 | 1 | 5.5 | 0.5 | 0 | 10 |
| torA-R9 101.8μg/mL | bufferH | MilliQ | EcoR1 | Pst1 | Total |
|---|---|---|---|---|---|
| 3 | 1 | 5.5 | 0 | 0.5 | 10 |
PCR
| torA-R9 | MilliQ | 10xPCR buffer for KOD plus neo | 2mM dNTP3 | 25mM MgSO4 | M13 | KOD plus neo | Total |
|---|---|---|---|---|---|---|---|
| 0.5 | 34 | 5 | 5 | 3 | 1.5 | 1 | 50 |
Predenature 94℃ 2min
Denature 98℃ 10sec
Annealing 60℃ 30sec
Extension 68℃ 20sec
→35cycles
Electrophoresis
torA-R9 plasmid, (EcoR1), (Pst1), (EcoR1,Pst1)
Lane1: 1kb ladder
Lane2: empty
Lane3: plasmid
Lane4: EcoR1
Lane5: Pst1
Lane6: EcoR1,Pst1
Lane7: empty
Lane8: 1kb ladder
Liquid culture
LacP-TatA-GFP-DT (9/4)
PCR purification
torA-R9 68.0μg/mL
September 20
Gel extraction
LacP-kil-DT (EcoR1,Pst1) (Xba1,Pst1)
Restrictive Digestion
| torA-R9 68.0μg/mL | bufferM | Spe1 | MilliQ | Total |
|---|---|---|---|---|
| 20 | 3 | 1 | 6 | 30 |
at 37℃, 2h
Ligation
| LacP-kil-DT(Xba1,Pst1) 12.4μg/mL | pSB1C3(EcoR1,Pst1) 34.2μg/mL | J23107-TatABCD-PsPA-DT(EcoR1,Spe1) 14.2μg/mL | Ligation High Ver.2 | Total |
|---|---|---|---|---|
| 7 | 4 | 7 | 9 | 27μL |
(Negative control)
| pSB1C3(EcoR1,Pst1) | Ligation High Ver.2 | MilliQ | Total |
|---|---|---|---|
| 4 | 9 | 14 | 27μL |
Transformation
Using above Ligation and Negative control's product and torA-R9.
Miniprep
J23106-kil-DT 6; 12.0μg/mL, (260/280): 1.78, (260/230): 1.73
J23106-kil-DT 7; 13.6μg/mL, (260/280): 1.85, (260/230): 1.73
Restrictive Digestion
| GFP-DT 84.1μg/mL | bufferM | 10xBSA | BbaI | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 3 | 3 | 1 | 8 | 30 |
at 37℃, 2 hours
Colony PCR
We made J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT's 1~8 samples.
| Quick tag | Tat f primer | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Predenature 94℃ 2min
Denature 94℃ 30sec
Annealing 54℃ 30sec
Extension 68℃ 4min
→30cycles
J23701-TatABCD-pspA-DT-LacP-torA-GFP-DT
Lane1: 1kb ladder
Lane2: 1
Lane3: 2
Lane4: 3
Lane5: 4
Lane6: 5
Lane7: 6
Lane8: 7
Lane9: 8
Lane10: 1kb ladder
Lane11: empty
Lane12: empty
→Bands didn't appear so we do PCR for 10 more cycles.
Liquid culture
We did liquid culture J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT's 1~4 samples.
Gel extraction
torA-R9 (EcoR1,Pst1): 31μg/mL, (280/280): 0.80, (280/230): 0.39
pSB1C3(EcoR1,Pst1): 22μg/mL, (280/280):1.33, (280/230):0.6
torA-R9 (Spe1): 56μg/mL, (280/280): 1.11, (280/230): 0.69
GFP-DT(Xba1): 45μg/mL, (280/280): 1.15, (280/230): 0.88
Ligation
| torA-R9 (EcoR1,Pst1) | pSB1C3(EcoR1,Pst1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 8 | 2 | 5 | 15μL |
| torA-R9 (Spe1) | pSB1C3(Xba1) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 3 | 3 | 3 | 9μL |
at 16℃, 1hour
Colony PCR
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (4k5)
numbering 9~15
Transformation
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (4k5)
J23107-TatABCD-pspA-DT-LacP-torA-kil-DT(pSB1C3)
Negative control (pSB1C3)
Negative control(pSB4K5)
torA-R9(pSB1C3)
Liquid culture
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT: colony PCR(9~15)
| 2xQuick tag | VF | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
Predenature 94℃ 2min
Denature 94℃ 30sec
Annealing 55℃ 30sec
Extension 68℃ 5min
→30cycles
PCR
| torA-R9-GFP-DT | MilliQ | KOD plus ver.2 buffer | dNTP3 | MgSO4 | M13 | VR | KOD plus | Total |
|---|---|---|---|---|---|---|---|---|
| 1 | 35 | 5 | 5 | 2 | 1.5 | 1.5 | 1 | 52 |
Predenature 94℃ 2min
Denature 94℃ 15sec
Annealing 55℃ 30sec
Extension 68℃ 5min
→30cycles
Miniprep
J23701-TatABCD-pspA-DT-LacP-torA-GFP-DT:2,3
2: 10.4μg/mL, (250/280): 1.45, (260/230): 0.95
3: 6.0μg/mL, (250/280): 1.32, (260/230): 0.79
(*)100μL
Electrophoresis
torA-R9-GFP-DT(after PCR)
September 21
Gel extraction
torA-R9-GFP-DT(after PCR): 10.3μg/mL, (260/280): 1.28, (220/230): 0.49

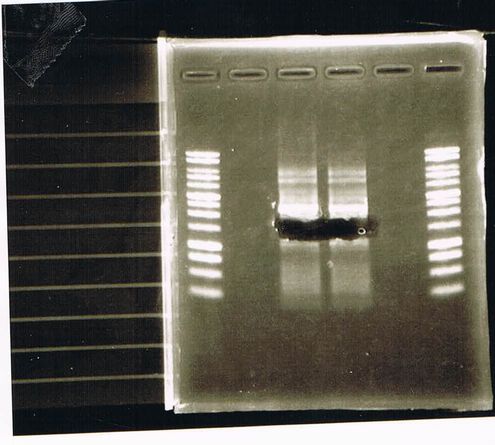
Electrophoresis
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(kan): 9~15
Electrophoresis
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT: 1,2
Lane1: empty
Lane2: empty
Lane3: 2
Lane4: 3
Lane5: 1kb ladder
Restrictive Digestion
| LacP-kil-DT(3) | Xba1 | Pst1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 7 | 3 | 30 |
| LacP-torA-GFP-DP(9/5) | Xba1 | Pst1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 7 | 3 | 30 |
| LacP-kil-DT | Xba1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|
| 2 | 0.5 | 1 | 5.5 | 1 | 10 |
| LacP-torA-GFP-DP | Xba1 | 10xBSA | MilliQ | BufferM | Total |
|---|---|---|---|---|---|
| 2 | 0.5 | 1 | 5.5 | 1 | 10 |
| LacP-kil-DT | Pst1 | MilliQ | BufferH | Total |
|---|---|---|---|---|
| 2 | 0.5 | 6.5 | 1 | 10 |
| LacP-torA-GFP-DP | Xba1 | Pst1 | MilliQ | BufferH | Total |
|---|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 1 | 10 |
| J23107-TatABCD-pspA-DT | Spe1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 20 | 1 | 1 | 4 | 14 | 40 |
| J23107-TatABCD-pspA-DT | Spe1 | BufferM | MilliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
| J23107-TatABCD-pspA-DT | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
at 37℃, 2hours
Electrophoresis
lane1. 1kb ladder
lane2. empty
lane3. LacP-kil-DT plasmid
lane4. LacP-kil-DT(Xba1)
lane5. LacP-kil-DT(Pst1)
lane6. LacP-kil-DT(Xba1, Pst1)
lane7. empty
lane8. LacP-torA-GFP-DP plasmid
lane9. LacP-torA-GFP-DP(Xba1)
lane10. LacP-torA-GFP-DP(Pst1)
lane11. LacP-torA-GFP-DP(Xba1, Pst1)
lane12. 1kb ladder
lane1. 1kb ladder
lane2. empty
lane3. J23107-TatABCD-pspA-DT plasmid
lane4. J23107-TatABCD-pspA-DT(Spe1)
lane5. J23107-TatABCD-pspA-DT(Pst1)
lane6. J23107-TatABCD-pspA-DT(Spe1, Pst1)
Colony PCR
- torA-R9(pSB1C3) (1-4)
| 10×Buffer | 2mM dNTPs | 25mM MgSO4 | VF | VR | KOD Plus Neo | MilliQ | Total |
|---|---|---|---|---|---|---|---|
| 5 | 5 | 3 | 1.5 | 1.5 | 1 | 33 | 50 |
94℃, 2min
98℃, 10sec
60℃, 30sec
68℃, 30sec
→30cycles
Electrophoresis
lane1. 1kb ladder
lane2. empty
lane3. torA-R9-1
lane4. torA-R9-2
lane5. torA-R9-3
lane6. torA-R9-4
lane7. empty
lane8. empty
September 22
Miniprep
torA-R9(Amp)1: 113.7μg/mL, (260/280): 1.96, (280/230): 2.42
torA-R9(Amp)2: 97.8μg/mL, (260/280): 1.67, (280/230): 1.72
torA-R9(Amp)3: 104.2μg/mL, (260/280): 1.93, (280/230): 2.67
LacP-torA-GFP-DT 6: 37.4μg/mL, (260/280): 2.24, (280/230): 3.27
Electrophoresis
LaneA: J23107-TatABCD (9/6)
LaneB: J23107-TatABCD-pspA-DT pSB3C5 (9/13)
LaneC: J23107-TatABCD-pspA-DT 1C5 (9/15)
LaneD: J23107-TatABCD-pspA-DT (9/15)
LaneE: J23107-TatABCD-pspA-DT(pSB1C3) (9/18)
LaneF: J23107-TatABCD-pspA-DT(pSB3C5) (9/18)
LaneG: J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:2 (9/20)
LaneH: J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:3 (9/20)
LaneA~H are (EcoR1,Pst1)
Restrictive Digestion
| J23107-TatABCD-pspA-DT-LacPtorA-GFP-DT:2 | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 15 | 3 | 1 | 12 | 30 |
| J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:3 | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 15 | 3 | 1 | 12 | 30 |
Restrictive Digestion
| LacP-torA-GFP-DT(37.4μg/mL) | BufferH | EcoR1 | Pst1 | Total |
|---|---|---|---|---|
| 25 | 3 | 1 | 1 | 30 |
Electrophoresis
①: J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:2
②: LacP-torA-GFP-DT(EcoR1,Pst1) (9/22)
③: J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT:3
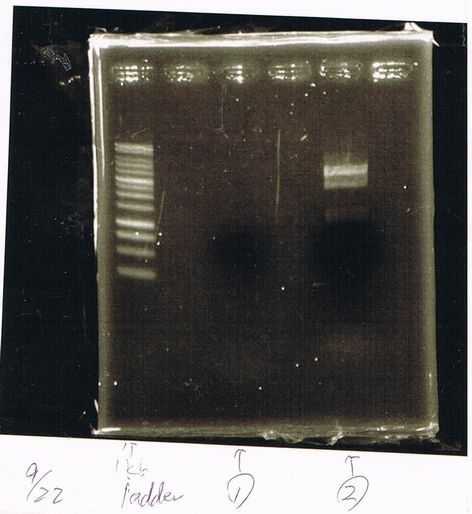
Gel extraction
LacP-torA-GFP-DT(EcoR1,Pst1) 5.5μg/ml 1.21(260/280) 0.37(260/230)
Purification
LacP-torA-GFP-DT 78.4μg/ml 1.62(260/280) 1.03(260/230)
Restrictive Digestion
| J23107-TatABCD-pspA-DT(pSB1C3) 78μg/ml | BufferH | Spe1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 3 | 1 | 1 | 10 | 30 |
| J23107-TatABCD-pspA-DT(pSB1C3) 78μg/ml | BufferH | Spe1 | MilliQ | Total |
|---|---|---|---|---|
| 3 | 1 | 0.5 | 5.5 | 10 |
| J23107-TatABCD-pspA-DT(pSB1C3) 78μg/ml | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 3 | 1 | 0.5 | 5.5 | 10 |
| LacP-torA-GFP-DT 78.4μg/ml | BufferM | BSA | Xba1 | Pst1 | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 3 | 3 | 1 | 1 | 7 | 30 |
| LacP-torA-GFP-DT 78.4μg/ml | BufferM | BSA | Xba1 | MilliQ | Total |
|---|---|---|---|---|---|
| 3 | 1 | 1 | 0.5 | 4.5 | 10 |
| LacP-torA-GFP-DT 78.4μg/ml | BufferH | Pst1 | MilliQ | Total |
|---|---|---|---|---|
| 3 | 1 | 0.5 | 5.5 | 10 |
at 37℃ for 2 hours
Electrophoresis
Lane1: 1kb ladder
Lane2: empty
Lane3: J23107-TatABCD-pspA-DT(pSB1C3)
Lane4: J23107-TatABCD-pspA-DT(Spe1, Pst1)
Lane5: J23107-TatABCD-pspA-DT(Spe1)
Lane6: J23107-TatABCD-pspA-DT(Pst1) (9/18)
Lane7: empty
Lane8: LacP-torA-GFP-DT
Lane9: LacP-torA-GFP-DT(Xba1, Pst1)
Lane10: LacP-torA-GFP-DT(Xba1)
Lane11: LacP-torA-GFP-DT(Pst1)
Lane12: empty
Lane13: 1kb ladder
September 23
Gel extraction
- LacP-torA-GFP-DT(EcoR1, Pst1)
lane1 1kb ladder
lane3 DNA
lane4 DNA
lane5 DNA
→failed
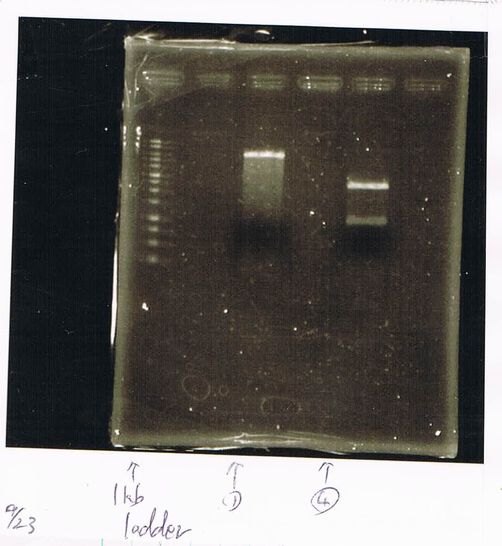

lane1 1kb ladder
lane3 J23107-TatABCD-pspA-DT(pSB1C3)(Spe1, Pst1)
lane5 LacP-torA-GFP-DT(Xba1, Pst1)
→failed
Restrictive Digestion
| LacP-torA-GFP-DT (78.4μg/mL) | Xba1 | Pst1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 3 | 7 | 30 |
| LacP-torA-GFP-DT (78.4μg/mL) | Xba1 | BufferM | BSA | MilliQ | Total |
|---|---|---|---|---|---|
| 3 | 0.5 | 1 | 1 | 4.5 | 10 |
| J23107-TatABCD-pspA-DT(pSB1C3) (78μg/mL) | Spe1 | Pst1 | BufferH | MilliQ | Total |
|---|---|---|---|---|---|
| 9 | 1 | 1 | 2 | 7 | 20 |
Ligation
| LacP-torA-GFP-DT(EcoR1, Pst1) (5.5μg/mL) | pSB4K5 (9.6μg/mL) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 8 | 4 | 6 | 18 |
at 16℃ for 1 hour
Liquid culture
- LacP-TatABCD-pspA-DT (2, 3, 5, 6)
Colony PCR
LacP-TatABCD-pspA-DT (1-6)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 4min
→30cycles
Electrophoresis
①, LacP-torA-GFP-DT
②, LacP-torA-GFP-DT(Xba1, Pst1)
③, LacP-torA-GFP-DT(Xba1)
④, LacP-torA-GFP-DT(Pst1)
⑤, J23107-TatABCD-pspA-DT(Spe1, Pst1)
Liquid culture
- LacP-torA-GFP-DT (4, 7)
- LacP-GFP generater (1-3)
- T7-6His-GFP-DT (1, 2)
Gel extraction
- LacP-torA-GFP-DT (Xba1, Pst1) 6.8μg/mL 1.27(260/280) 0.40(260/230)
Purification
- J23107-TatABCD-pspA-DT(Spe1, Pst1) 15.1μg/mL 1.49(260/280) 0.80(260/230)
Ligation
| J23107-TatABCD-pspA-DT(Spe1, Pst1) (15.1μg/mL) | LacP-torA-GFP-DT (Xba1, Pst1) (6.8μg/mL) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 10 | 4 | 7 | 21 |
| J23107-TatABCD-pspA-DT(Spe1, Pst1) (15.1μg/mL) | LacP-kil-DT (Xba1, Pst1) (12.4μg/mL) | Ligation High Ver.2 | Total |
|---|---|---|---|
| 8 | 2 | 5 | 15 |
at 16℃ for 1 hour
Transformation
LacP-torA-GFP-DT(pSB4K5)
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT
J23107-TatABCD-pspA-DT-LacP-kil-DT
| DNA | competent cell | Total |
|---|---|---|
| 2 | 10 | 12 |
preculture at 37℃ for 1 hour culture at 37℃ for 16 hours
Electrophoresis
J23107-TatABCD-pspA-DT(pSB1C3) (1-6)
lane1 1kb ladder
lane2~7 DNA
lane8 1kb ladder
September 24
Miniprep
- J23107-TatABCD-pspA-DT-2 139.4μg/ml 1.69(260/280) 1.83(260/230)
- J23107-TatABCD-pspA-DT-3 134.7μg/ml 1.61(260/280) 1.69(260/230)
- J23107-TatABCD-pspA-DT-5 139.3μg/ml 1.55(260/280) 1.45(260/230)
- J23107-TatABCD-pspA-DT-6 138.5μg/ml 1.58(260/280) 1.40(260/230)
- LacP-torA-GFP-DT-4
- LacP-torA-GFP-DT-7
- T7-6His-GFP-DT-1
- T7-6His-GFP-DT-2
Colony PCR
LacP-GFP generater (1-3)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 30sec
55℃, 30sec
68℃, 1min
→30cycles
①-③, LacP-GFP generater (1-3)
Liquid culture
- J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (1-4)
- J23107-TatABCD-pspA-DT-LacP-kil-DT (1-3)
Colony PCR
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT (1-4)
J23107-TatABCD-pspA-DT-LacP-kil-DT (1-3)
| 2×Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
94℃, 2min
94℃, 30sec
55℃, 30sec
68℃, 5min
→30cycles
Electrophoresis
lane1, 1kb ladder
lane2-5, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (1-4)
lane6-8, J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (1-3)
September 25
Liquid culture
- J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB4K5)-2
Miniprep
J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3)-3: 26.2μg/mL, (260/280): 1.50, (280/230): 0.65
J23107-TatABCD-pspA-DT-LacP-torA-GFP(pSB1C3)-3: 24.8μg/mL, (260/280): 1.56, (280/230): 0.76
LacP-GFP generater-1: 42.0μg/mL, (260/280): 1.58, (280/230): 0.94
Restrictive Digestion
| J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (26.2ng/µL) | EcoRl | Spel | buffer M | MiliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
| J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (26.2ng/µL) | EcoRl | buffer M | MiliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
| J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (26.2ng/µL) | Spel | buffer M | MiliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
| J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (24.8ng/µL) | EcoRl | Pstl | buffer H | MiliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
| J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (24.8ng/µL) | Pstl | buffer H | MiliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
| J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (24.8ng/µL) | EcoRl | buffer H | MiliQ | Total |
|---|---|---|---|---|
| 2 | 0.5 | 1 | 6.5 | 10 |
at 37℃ for 2 hours
Electrophoresis
lane1, 1kb ladder
lane2, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3)
lane3, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (EcoRl, Spel)
lane4, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (EcoRl)
lane5, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (Spel)
lane6, J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3)
lane7, J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (EcoRl, Pstl)
lane8, J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (EcoRl)
lane9, J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (Pstl)
lane10, LacP-torA-GFP-DT(EcoR1,Xba1)(9/15)
lane11, LacP-torA-GFP-DT(EcoR1,Xba1)(9/17)
lane12, 1kb ladder
→The bands didn't appear, so we did one more electrophoresis.
lane1, 1kb ladder
lane2, LacP-torA-GFP-DT(EcoR1,Xba1)(9/17)
lane3, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3)
lane4, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (EcoRl, Spel)
lane5, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (EcoRl)
lane6, J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (Spel)
Liquid culture
- LacP-torA-GFP-DT(pSB4K5) (1-4)
Restrictive Digestion
| J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) | EcoRl | Spel | buffer M | MiliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
| J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) | EcoRl | Pstl | buffer H | MiliQ | Total |
|---|---|---|---|---|---|
| 15 | 1 | 1 | 3 | 10 | 30 |
at 37℃ for 1 hour
Electrophoresis
J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) (EcoRl, Spel)
J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3) (EcoRl, Pstl)
→The bands didn't appear.
Observation through a confocal microscope
① J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) ② J23107-TatABCD-pspA-DT-LacP-torA-GFP-DT(pSB1C3) ③ J23107-TatABCD-pspA-DT-LacP-kil-DT(pSB1C3)
→We observed nothing.
October 12
Transformation
| conpetent cell(3/15) | J23107-TatABCD-PspA-DT (9/13) (pSB3C5) | J23107-TatABCD-PspA-DT(9/18)(pSB3C5) | LacP-torA-GFP-DT(9/10) | total |
|---|---|---|---|---|
| 20μL | 2μL | 22μL | ||
| 20μL | 2μL | 22μL | ||
| 20μL | 2μL | 22μL |
preculture in SOC medium for 1 hour at 25 degree
Checking Colony PCR(negative control)
| Quick Taq | VF | VR | MilliQ | total |
|---|---|---|---|---|
| 25μL | 1μL | 1μL | 23μL | 50μL |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 5min
30 cycles
->Electrophoresis
October 15
Restrictive Digestion & Electrophoresis
| pSB4C5 | Xba1 | Pst1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10μL | 1μL | 1μL | 3μL | 3μL | 12μL | 30μL |
at 37℃ for 1 hour
Transformation
retry of lab work that we did on 10/12
| J23107-TatABCD-PspA-DT | LacP-torA-GFP-DT | competent cell | total |
|---|---|---|---|
| 2μL | 20μL | 22μL | |
| 2μL | 20μL | 22μL |
preculture in 100μL of SOC medium
October 16
Colony PCR
- 4 colonies of J23107-TatABCD-PspA-DT(9/18)
| Quick Taq | VF2 | VR | MilliQ | total |
|---|---|---|---|---|
| 25μL | 1μL | 1μL | 23μL | 50μL |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 4min
30 cycles
- 4 colonies of LacP-torA-GFP-DT(10/15)
| Quick Taq | VF2 | VR | MilliQ | total |
|---|---|---|---|---|
| 25μL | 1μL | 1μL | 23μL | 50μL |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 1.5min
30 cycles
October 17
Electrophoresis
product of colony PCR on 10/16
- fig.1
lane_1 : 1kb ladder
lane_2-5 : J23107-TatABCD-PspA-DT
lane_6-9 : LacP-torA-GFP-DT
lane_10 : 1kb ladder
we retried this because something is wrong with 1 kb ladder
- fig.2
lane_1 : 1kb ladder
lane_2 : J23107-TatABCD-PspA-DT(lane_3)
Liquid culture
J23017-TatABCD-PspA-DT(10/15)
Colony PCR
- 8 colonies of LacP-torA-GFP-DT(10/15)
| Quick Taq | VF2 | VR | MilliQ | total |
|---|---|---|---|---|
| 25μL | 1μL | 1μL | 23μL | 50μL |
94℃ 2min
94℃ 30sec
55℃ 30sec
68℃ 1.5min
30 cycles
->Electrophoresis
we named 8 colonies [5]~[12] because on 10/16, we did colony PCR using 4 colonies([1]~[4]) of J23107-TatABCD-PspA-DT.
- fig.3
lane_1 : 1kb ladder
lane_2 : [5]
lane_3 : [6]
lane_4 : [7]
lane_5 : [8]
- fig.4
lane_1 : 1kb ladder
lane_2 : [9]
lane_3 : [10]
lane_4 : [11]
lane_5 : [12]
Liquid culture
[5] and [7]
October 18
Miniprep
| gene | ng/μL |
|---|---|
| J23107-TatABCD-PspA-DT (10/17) | 134.0 |
| LacP-torA-GFP-DT [5] (10/17) | 94.7 |
October 19
Restrictive Digestion
- sample_1
| J23107-TatABCD-PspA-DT | EcoR1 | Spe1 | BufferM | MilliQ | total |
|---|---|---|---|---|---|
| 15μL | 1μL | 1μL | 3μL | 10μL | 30μL |
- sample_2
| LacP-torA-GFP-DT | Xba1 | Pst1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 10μL | 1μL | 1μL | 3μL | 3μL | 12μL | 30μL |
- sample_3
| LacP-kil-DT | Xba1 | Pst1 | BSA | BufferM | MilliQ | total |
|---|---|---|---|---|---|---|
| 12μL | 1μL | 1μL | 3μL | 3μL | 10μL | 30μL |
Electrophoresis
lane_1 : 1kb ladder
lane_2 : J23107-TatABCD-PspA-DT
lane_3 : LacP-torA-GFP-DT
lane_4 : LacP-kil-DT
Restrictive Digestion
| T7 | Spe1 | Pst1 | BufferH | MilliQ | total |
|---|---|---|---|---|---|
| 10μL | 1μL | 1μL | 2μL | 6μL | 20μL |
at 37℃ for 1 hour
Eledtrophoresis
lane_1 : 1kb ladder
lane_2 : sample 1
lane_3 : blank
lane_4 : sample 2
lane_5 : blank
lane_6 : sample 3
Though this figure did not indicate, there was a 1000 bp band that seems to be product of LacP-Kil-DT
October 22
Gel extraction
- LacP-torA-GFP-DT
- LacP-kil-DT
| Gene | ng/µL | 260/280 | 260/230 |
|---|---|---|---|
| LacP-torA-GFP-DT | 4,4 | 2,02 | 0,22 |
| LacP-kil-DT | 3,8 | 1,54 | 0,11 |
Restrictive Digestion
| LacP-torA-GFP-DT(pSB3C5, 9/10) | EcoR1 | Pst1 | Buffer M | MilliQ | Total |
|---|---|---|---|---|---|
| 10µL | 1µL | 1µL | 2µL | 6µL | 20µL |
At 37℃ for 1 hour
Electrophoresis
- fig.1
Lane_1:1kb ladder
Lane_2:empty
Lane_3:T7(S,P)
- fig.2
Lane_1:1kb ladder
Lane_2:LacP-torA-GFP-DT(E,P)
Gel Extraction
- fig.3, 4
Lane_1:1kb ladder
Lane_2:blank
Lane_3:T7(S,P)
| Gene | ng/µL | 260/280 | 260/230 |
|---|---|---|---|
| T7(S,P) | 5,4 | 1,82 | 0,23 |
- fig.5, 6
Lane_1:1kb ladder
Lane_2: LacP-torA-GFP-DT(E,P)
Lane_3:pSB4C5(X,P)
| Gene | ng/µL | 260/280 | 260/230 |
|---|---|---|---|
| LacP-torA-GFP-DT(E,P) | 4,9 | 1,66 | 0,28 |
| pSB4C5 | 23,9 | 1,88 | 0,88 |
Ligation
| LacP-torA-GFP-DT(X,P) | T7(S,P) | Ligation high Ver.2 | Total |
|---|---|---|---|
| 10µL | 2µL | 5,5µL | 17,5µL |
| LacP-torA-GFP-DT(E,P) | pSB4K5(E,P) | Ligation high Ver.2 | Total |
|---|---|---|---|
| 10µL | 2µL | 5,5µL | 17,5µL |
| LacP-torA-GFP-DT(X,P) | pSB4C5(X,P) | Ligation high Ver.2 | Total |
|---|---|---|---|
| 10µL | 2µL | 5,5µL | 17,5µL |
At 4℃ overnight
October 23
Transformation
- T7-LacP-kil-DT
- LacP-torA-GFP-DT(pSB4K5)
- LacP-torA-GFP-DT(pSB4C5)
transformed competent cells were plated on two plates respectively.(plate a and plate b)
Ligation
| pSB4C5(X,P) | LacP-GFP-DT(X,P) | Ligation high Ver.2 | Total |
|---|---|---|---|
| 2µL | 10µL | 6µL | 18µL |
At 4℃ overnight
October 24
Colony PCR
- T7-LacP-kil-DT(a1~a8)
- LacP-torA-GFP-DT(pSB4C5)(a1~a4,b1~b4)
- LacP-torA-GFP-DT(pSB4K5)(a1~a4,b1,b2)
| Quick Taq | VF2 | VR | MilliQ | Total |
|---|---|---|---|---|
| 25µL | 1µL | 1µL | 23µL | 50µL |
94℃ 2min
94 30sec
55 30sec
68 1min
30cycles
Transformation
- LacP-GFP-DT(pSB4C5)
Electrophoresis
Products of colony PCR
- fig.1
Lane_1 :1kb ladder
Lane_2 :empty
Lane_3 : T7-LacP-kil-DT(a1)
Lane_4 : T7-LacP-kil-DT(a2)
Lane_5 : LacP-torA-GFP-DT(pSB4C5)(a1)
Lane_6 : LacP-torA-GFP-DT(pSB4C5)(a2)
Lane_7 : LacP-torA-GFP-DT(pSB4C5)(b1)
Lane_8 : LacP-torA-GFP-DT(pSB4C5)(b2)
Lane_9 : LacP-torA-GFP-DT(pSB4K5)(a1)
Lane_10 : LacP-torA-GFP-DT(pSB4K5)(a2)
Lane_11 : LacP-torA-GFP-DT(pSB4K5)(b1)
Lane_12 : LacP-torA-GFP-DT(pSB4K5)(b2)
- fig.2
Lane_1 :1kb ladder
Lane_2 : T7-LacP-kil-DT(a3)
Lane_3 : T7-LacP-kil-DT(a4)
Lane_4 : T7-LacP-kil-DT(a5)
Lane_5 : T7-LacP-kil-DT(a6)
Lane_6 : T7-LacP-kil-DT(a7)
Lane_7 : T7-LacP-kil-DT(a8)
Lane_8 : LacP-torA-GFP-DT(pSB4K5)(a3)
Lane_9 : LacP-torA-GFP-DT(pSB4K5)(a4)
Lane_10 :1kb ladder
Liquid culture
- LacP-torA-GFP-DT(pSB4C5)(a1,a2)
- T7-LacP-kil-DT(a6)
October 25
Observation through a confocal microscope
- LacP-torA-GFP-DT(pSB4C5)
We observed nothing.
September 6
PCR and Electrophoresis assay
①psB1K3 ②lacI ③GFP ④GFP ⑤RFP ⑥RFP ⑦CFP ⑧DT
| Quick Taq | primer F | Primer R | DNA(①-⑧) | milliQ | total |
|---|---|---|---|---|---|
| 12.5 | 0.5 | 0.5 | 1 | 10.5 | 25 |
94℃ 2min, (94℃ 30sec, 50℃ 30sec)x25cycles, 68℃ 50sec
September 7
PCR and Electrophoresis assay of psB1K3
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(psB1K3) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec)x30cycles, 68℃ 2min
September 10
PCR and Electrophoresis assay of CFP
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(CFP) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 48℃ 30sec)x30cycles, 68℃ 45sec
PCR and Electrophoresis assay of lacI
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(lacI) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec)x30cycles, 68℃ 30sec
PCR
①PCR product of psB1K3 ②psB1K3 ③psB1C3 ④psB1C3 ⑤PCR product of CFP
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(①-⑤) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec)x30cycles, 68℃ 2min20sec
September 11
PCR and Electrophoresis assay
①DT ②PCR product of DT ③PCR product of CFP ④CFP(2011 plate)
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(①-④) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 48℃ 30sec)x30cycles, 68℃ 1min
PCR and Electrophoresis assay
①DT 92.1ng/µl ②DT 28.0ng/µl ③DT
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(①-③) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 50℃ 30sec)x30cycles, 68℃ 30sec
September 12
Purification of PCR product
psB1K3(9/10④) 55µg/ml lacI(9/10) 90µg/ml GFP(9/6) 42µg/ml RFP(9/6) 71µg/ml DT(9/11) 81µg/ml GFP 37µg/ml RFP 57µg/ml
Golden Gate Assembly
| psB1K3 | lacI | GFP(2'-3) | RFP(3'-5) | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|
| 1.8 | 0.1 | 1.0 | 0.6 | 0.1 | 1.5 | 0.15 | 1.0 | 1.0 | 8.15 | 15 |
| psB1K3 | lacI | GFP(2'-5) | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|
| 1.8 | 0.1 | 1.0 | 0.1 | 1.5 | 0.15 | 1.0 | 1.0 | 8.75 | 15 |
| psB1K3 | lacI | GFP(2'-3) | RFP(3'-5) | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 1.0 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 6.95 | 15 |
| psB1K3 | lacI | GFP(2'-3) | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.95 | 15 |
Transformation
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
September 13
Transformation
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
September 14
Liquid Culture
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
September 15
Miniprep
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
①4.0 µg/ml ②17 µg/ml ③15 µg/ml ④8.0 µg/ml
Restriction Enzyme Digestion
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
| DNA(①-④) | 10x NEB Bufer3 | BSA | EcoR1 | MilliQ | Total |
|---|---|---|---|---|---|
| 9 | 2 | 0.5 | 0.5 | 8 | 20 |
September 17
Electrophoresis assay
①lacI-GFP(2'-3)-RFP(3'-5)-DT
②lacI-GFP(2'-5)-DT
③lacI-GFP(2'-3)-RFP(3'-5)-DT
④lacI-GFP(2'-5)-DT
September 18
Restriction Enzyme Digestion
③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
| DNA(③,④) | 10x H Bufer | BSA | EcoR1 | MilliQ | Total |
|---|---|---|---|---|---|
| 15 | 3 | 0.5 | 0.5 | 11 | 30 |
Colony PCR
①lacI-GFP(2'-3)-RFP(3'-5)-DT ②lacI-GFP(2'-5)-DT ③lacI-GFP(2'-3)-RFP(3'-5)-DT ④lacI-GFP(2'-5)-DT
| 2x Quick Tag Dye Mix | VF primer | VR primer | MiliQ | Total |
|---|---|---|---|---|
| 25 | 1 | 1 | 23 | 50 |
September 19
DpnI Restriction Enzyme Digestion of psB1K3
| psB1K3 | DpnI | Total |
|---|---|---|
| 10 | 0.2 | 10.2 |
37℃ 1h incubate
Ligation of psB1K3
| psB1K3 | Ligation High | MiliQ | TH Kinase | Total |
|---|---|---|---|---|
| 2 | 5 | 7 | 7 | 15 |
16.0℃ 1.5h
September 20
Transformation of psB1K3
September 21
PCR of psB3C5 and Electrophoresis assay
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(psB3C5) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec)x30cycles, 68℃ 2min
PCR and Electrophoresis assay
①psB3C5 ②psB1K3
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(①,②) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5.0 | 5.0 | 2.0 | 1.5 | 1.5 | 1.0 | 1.0 | 33 | 50 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec, 68℃ 2min)x30cycles
PCR of DT and Electrophoresis assay
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA(DT) | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5.0 | 5.0 | 2.0 | 1.5 | 1.5 | 1.0 | 1.0 | 33 | 50 |
94℃ 2min, (94℃ 15sec, 50℃ 30sec, 68℃ 30sec)x30cycles
September 22
Colony PCR (Colony plated in September 20)
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 5.0 | 5.0 | 2.0 | 1.5 | 1.5 | 1.0 | 1.0 | 33 | 50 |
94℃ 2min, (94℃ 15sec, 58℃ 30sec, 68℃ 2min20sec)x25cycles
PCR of Primers and Electrophoresis assay
①J23106(1'-2) ②J23106(2'-3) ③J23106(3'-4) ④J23106(4'-5) ⑤J23106(3'-5) ⑥J23106(2'-5) ⑦J23106(1'-5) ⑧araC(1'-6) ⑨araC(5'-6)
| 10x Buffer KOD plus | 2mM dNTPs | 25mM MgSO4 | F primer | R primer | DNA | KOD plus | milliQ | total |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 1.0 | 0.75 | 0.75 | 0.5 | 0.5 | 16.5 | 25 |
94℃ 2min, (94℃ 15sec, 56℃ 30sec)x30cycles, 68℃ 20sec
①J23106(1'-2) 60µg/ml ②J23106(2'-3) 63µg/ml ③J23106(3'-4) 48µg/ml ④J23106(4'-5) 60µg/ml ⑤J23106(3'-5) 62µg/ml ⑥J23106(2'-5) 40µg/ml ⑦J23106(1'-5) 29µg/ml ⑧araC(1'-6) 60µg/ml ⑨araC(5'-6) 62µg/ml
Ligation
4 promoters
| psB3C5 | ①J23106 (1'-2) | ②J23106 (2'-3) | ③J23106 (3'-4) | ④J23106 (4'-5) | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 2.0 | 2.0 | 2.5 | 2.0 | 2.0 | 3.0 | 3.0 | 1.5 | 0.15 | 1.0 | 1.0 | 7.85 | 30 |
3 promoters
| psB3C5 | ①J23106 (1'-2) | ②J23106 (2'-3) | ⑤J23106 (3'-5) | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 2.0 | 2.0 | 2.5 | 2.0 | 3.0 | 3.0 | 1.5 | 0.15 | 1.0 | 1.0 | 9.85 | 30 |
2 promoters
| psB3C5 | ①J23106(1'-2) | ⑥J23106(2'-5) | ⑨araC(5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 2.0 | 2.5 | 2.0 | 3.0 | 3.0 | 1.5 | 0.15 | 1.0 | 11.85 | 30 |
1 promoter
| psB3C5 | ⑦J23106(1'-5) | ⑨araC(5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 2.5 | 2.0 | 3.0 | 3.0 | 1.5 | 0.15 | 1.0 | 1.0 | 13.85 | 30 |
0 promoter
| psB3C5 | ⑨araC(5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 2.0 | 3.0 | 3.0 | 1.5 | 0.15 | 1.0 | 1.0 | 16.35 | 30 |
September 23
Transformation of product of ligation(9/22)
September 24
Transformation of product of ligation(9/22)
September 25
Liquid Culture
colony1,2,3 of plate2,3
Ligation
4 promoters
| psB3C5 | ① J23106 (1'-2) | ② J23106 (2'-3) | ③ J23106 (3'-4) | ④ J23106 (4'-5) | ⑨ araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.05 | 15 |
3 promoters
| psB3C5 | ①J23106 (1'-2) | ②J23106 (2'-3) | ⑤J23106 (3'-5) | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 0.3 | 0.2 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.25 | 15 |
2 promoters
| psB3C5 | ①J23106 (1'-2) | ⑥J23106 (2'-5) | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 0.3 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.45 | 15 |
1 promoter
| psB3C5 | ⑦J23106 (1'-5) | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.6 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.35 | 15 |
0 promoter
| psB3C5 | ⑨araC (5'-6) | GFP | DT | 10x NEB T4 Buffer | 100x BSA | BsaI | NEB T4 ligase | milliQ | total |
|---|---|---|---|---|---|---|---|---|---|
| 2.0 | 0.2 | 1.0 | 0.2 | 1.5 | 0.15 | 1.0 | 1.0 | 7.95 | 15 |
(37℃ 3min, 16℃ 4min)x25cycles,
 "
"