Team:HokkaidoU Japan/Project/PHB Synthesis
From 2012.igem.org
(→Construction of P(3HB) producing module) |
(→Discussion) |
||
| (One intermediate revision not shown) | |||
| Line 41: | Line 41: | ||
==Discussion== | ==Discussion== | ||
<div class="hokkaidou-section"> | <div class="hokkaidou-section"> | ||
| - | <p>We succeeded in optimization of plastic production using pGEM(phaCAB), however, pSB(phaCAB) did not work as we expected. JM109 is more powerful than DH5α because it is closely related to wild type. Pantothenic acid and glucose are substrates of P(3HB) synthesis pathway, so the result is quite natural. TB medium includes phosphate buffer. It keeps pH of culture fluid as neutral, so bacteria can grow | + | <p>We succeeded in optimization of plastic production using pGEM(phaCAB), however, pSB(phaCAB) did not work as we expected. JM109 is more powerful than DH5α because it is closely related to wild type. Pantothenic acid and glucose are substrates of P(3HB) synthesis pathway, so the result is quite natural. TB medium includes phosphate buffer. It keeps pH of culture fluid as neutral, so bacteria can grow and produce polymer more efficiently. ''E. coli'' which was transformed with pGEM(phaCAB) produced plenty of PHB, so that our method of culturing and producing PHB works well. As for the reason why PHB was not produced with our phaCAB BioBrick part, we think that an over expression of phaCAB genes caused the resulting enzymes being transferred into inclusion bodies. We assure that ''E. coli'' which is transformed pGEM(phaCAB) produces P(3HB) because pGEM(phaCAB) includes phaCAB operon which derived from ''R. eutropha'' genome. However, in our experiment, we used PhaC (BBa_K342001, INSA-Lyon2010) which has codon optimization to be fit for translation of ''E. coli''. Furthermore, RBS and promoter which we used are ''E. coli'' genes. TetR repressible promoter (BBa_R0040 ) is stronger than a promoter on pGEM(phaCAB). In this case, phaCAB genes may be expressed more in ''E. coli'' than in the case of pGEM(phaCAB).</p> |
<p>To confirm our hypothesis, we should examine whether the intermediates are produced. The data that two intermediates of P(3HB) producing pathway are synthesized but not the final product should mean that the enzymes are put in inclusion bodies. In addition, to make our phaCAB BioBrick part better we will replace TetR repressible promoter with pGEM(phaCAB) promoter. We guess that due to the pGEM(phaCAB) promoter the expression of phaCAB genes will be less than before, and PHB is produced successfully.</p> | <p>To confirm our hypothesis, we should examine whether the intermediates are produced. The data that two intermediates of P(3HB) producing pathway are synthesized but not the final product should mean that the enzymes are put in inclusion bodies. In addition, to make our phaCAB BioBrick part better we will replace TetR repressible promoter with pGEM(phaCAB) promoter. We guess that due to the pGEM(phaCAB) promoter the expression of phaCAB genes will be less than before, and PHB is produced successfully.</p> | ||
<p>We hope many iGEM teams will be able to produce plastic easily with our parts and they will make future iGEM projects more eco-friendly and more attractive.</p> | <p>We hope many iGEM teams will be able to produce plastic easily with our parts and they will make future iGEM projects more eco-friendly and more attractive.</p> | ||
Latest revision as of 04:06, 27 September 2012
Contents |
Planning
We attempted to make Escherichia coli producing bio-plastic for demonstrating industrial applicability of “Aggregation Module” that makes “bio-capsules” formed by aggregation of E. coli through cell-cell interaction via adhesion molecule, Ag43, located on the surface of outer membrane.
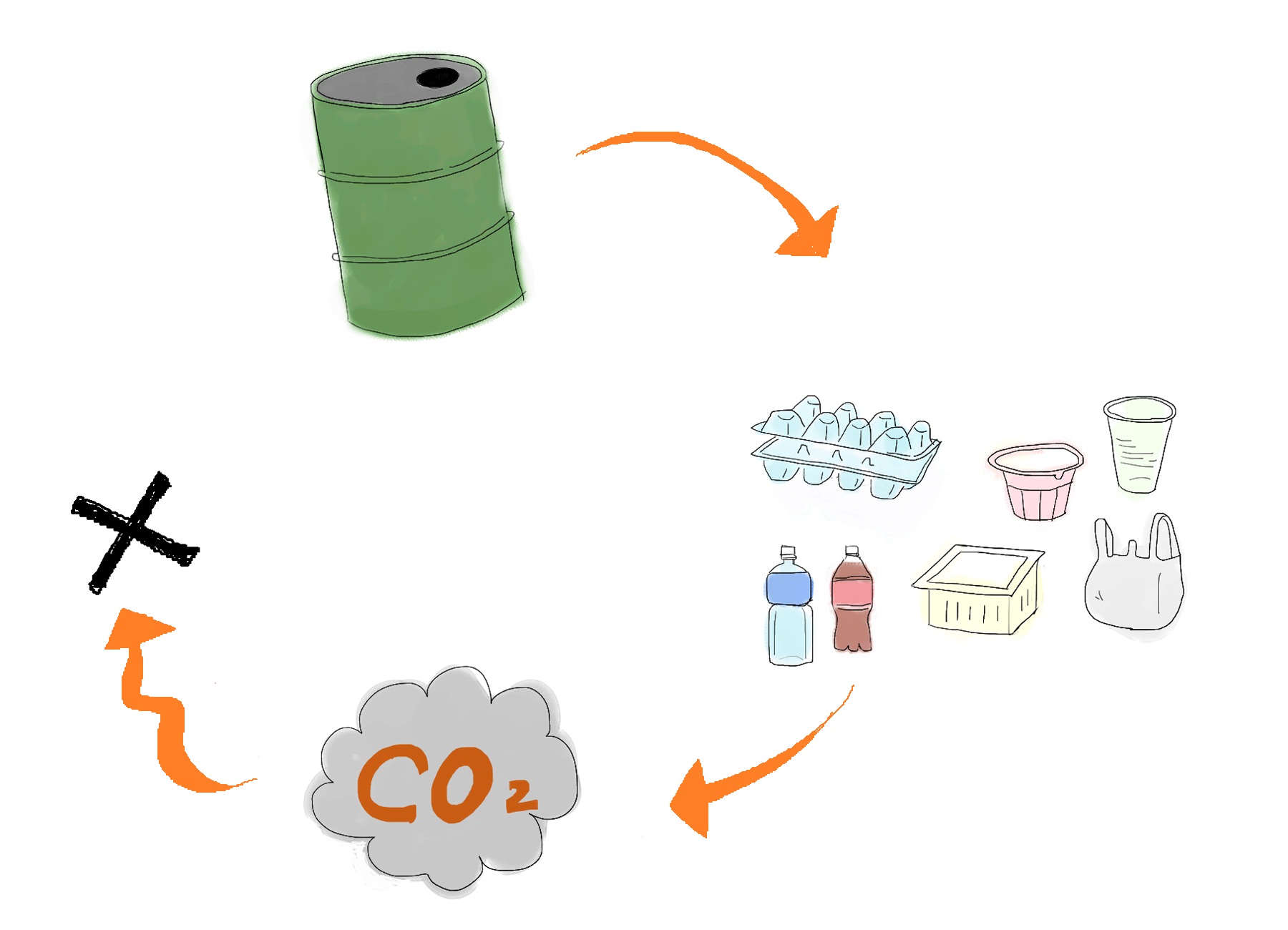
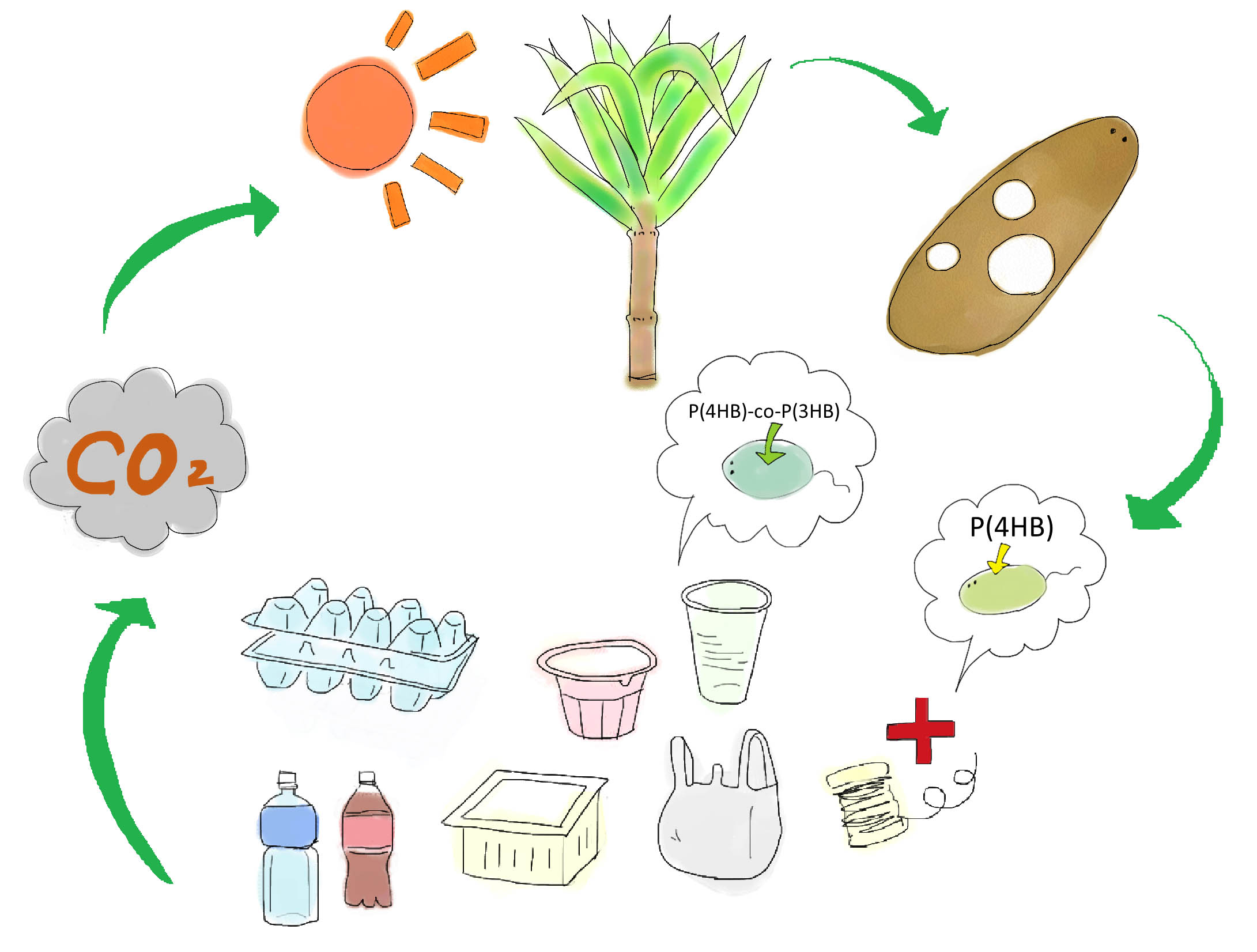
Development of a cost-effective method for manufacturing biodegradable plastic is one of important issues for making sustainable future society. Common plastic is made from oil (Fig. 1). Oil is limited resource, since it is estimated to be exhausted just in 46.2 years[1]. Synthetic plastic also cause problem as a stable waste material since it is not bio-degradable. Bio-Plastic will never contribute to the increase of atmospheric carbon dioxide since bacteria utilizes glucose as resource for its biosynthesis (Fig. 2).
Background
Poly-3-hydroxy-butyrate P(3HB) is one of bio-plastics made by bacteria and archaea. They store P(3HB) in their cells as energy-storage molecule[2]. So bio-plastic is bio-degradable. Those bacteria synthesize it from acetyl-CoA, the intermediate product of glycolysis. Monomer of P(3HB) is also used as material for making several other useful plastics (co-polymer) through coupling with other types of component molecules like valeric acid or lactic acid[3]. Ralstonia eutropha is well-known as hydrogen bacteria that produce P(3HB).
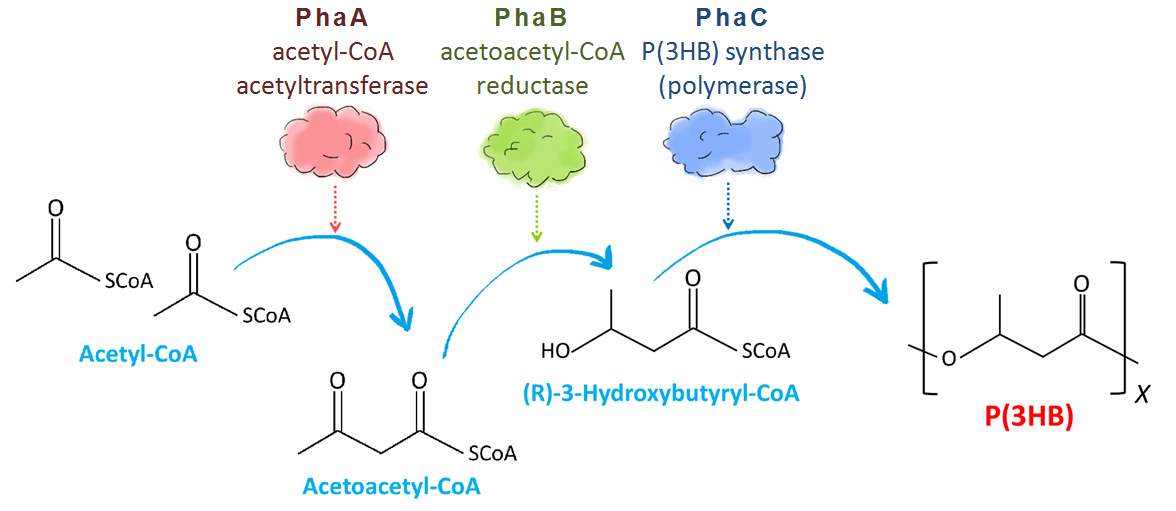
In R. eutropha cells, P(3HB) is made through 3 steps (Fig. 3). Two acetyl-CoA molecules made from carbohydrate converted to acetoacetyl-CoA by acetyl-CoA acetyltransferase encoded by PhaA gene. Then acetoacetyl-CoA reductase encoded by PhaB gene catalyzes the reduction of acetoacetyl-CoA to (R)-3-hydroxybutyryl-CoA (3HB-CoA). Finally, P(3HB) synthase encoded by PhaC gene catalyzes polymerization reaction of monomer molecules to polymer P(3HB)[2].
Experiments
Optimization of P(3HB) production
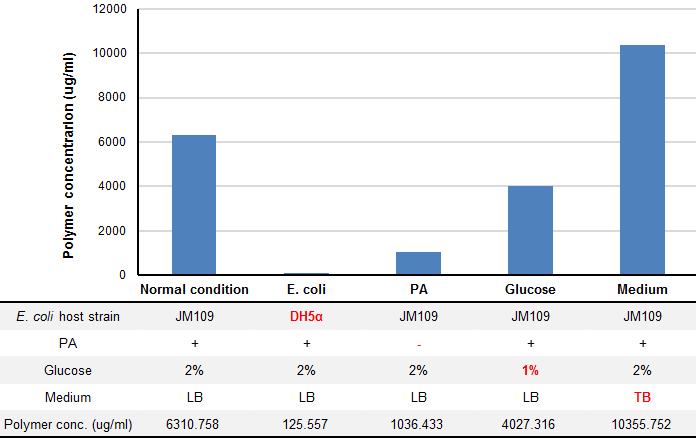
We tried optimization of P(3HB) production under four conditions: E. coli host strain, addition of pantothenic acid (PA), Glucose concentration and liquid culture mediun. P(3HB) wad produced by using pGEM(phaCAB) that covers all region need for P(3HB) biosynthesis found in R. eutropha genome, which was provided by Taguchi Lab.
Construction of P(3HB) producing module
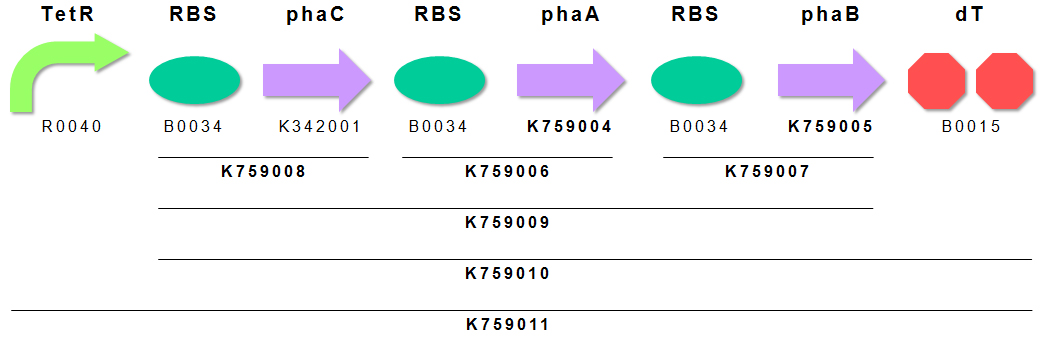
We constructed P(3HB) producing module K759011 (Fig. 4). Genes encoding PhaA and PhaB were obtained by subcloning from pGEM(phaCAB). PhaC has already registered as BBa_K342001 by INSA-Lyon2010. We appreciate INSA-Lyon 2010 teams effort. We ligated these genes in the order of PhaC, PhaA, PhaB for making BioBrick of “Plastic Producing Module”. TetR repressible promoter (BBa_R0040 ) and RBS (BBa_B0034) are fused to upstream of above gene cluster.
Results
Optimization of P(3HB) production
P(3HB) production was drastically changed in several conditions (Fig. 5). “Plastic producing module” in JM109 have shown more efficient plastic production than that in DH5α (1.45 fold). In the same condition, concentration of glucose and presence of PA affects the yield of plastic production. We found that TB medium, rich medium, produced 1.64 fold more plastic than LB medium.
Construction of P(3HB) producing module
We did not succeed in producing plastic by culturing JM109/pSB(phaCAB) containing BioBrick part in pSB1C3 yet. Now we are checking several culture condition and promoter species for controlling their gene expression.
Discussion
We succeeded in optimization of plastic production using pGEM(phaCAB), however, pSB(phaCAB) did not work as we expected. JM109 is more powerful than DH5α because it is closely related to wild type. Pantothenic acid and glucose are substrates of P(3HB) synthesis pathway, so the result is quite natural. TB medium includes phosphate buffer. It keeps pH of culture fluid as neutral, so bacteria can grow and produce polymer more efficiently. E. coli which was transformed with pGEM(phaCAB) produced plenty of PHB, so that our method of culturing and producing PHB works well. As for the reason why PHB was not produced with our phaCAB BioBrick part, we think that an over expression of phaCAB genes caused the resulting enzymes being transferred into inclusion bodies. We assure that E. coli which is transformed pGEM(phaCAB) produces P(3HB) because pGEM(phaCAB) includes phaCAB operon which derived from R. eutropha genome. However, in our experiment, we used PhaC (BBa_K342001, INSA-Lyon2010) which has codon optimization to be fit for translation of E. coli. Furthermore, RBS and promoter which we used are E. coli genes. TetR repressible promoter (BBa_R0040 ) is stronger than a promoter on pGEM(phaCAB). In this case, phaCAB genes may be expressed more in E. coli than in the case of pGEM(phaCAB).
To confirm our hypothesis, we should examine whether the intermediates are produced. The data that two intermediates of P(3HB) producing pathway are synthesized but not the final product should mean that the enzymes are put in inclusion bodies. In addition, to make our phaCAB BioBrick part better we will replace TetR repressible promoter with pGEM(phaCAB) promoter. We guess that due to the pGEM(phaCAB) promoter the expression of phaCAB genes will be less than before, and PHB is produced successfully.
We hope many iGEM teams will be able to produce plastic easily with our parts and they will make future iGEM projects more eco-friendly and more attractive.
References
- H.Ohara. Change from Oil-based to Bio-based. Sen’i gakkaishi 66, 4.129-132 (2010)
- Pohlmann, A. et al. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nature biotechnology 24, 1257–62 (2006).
- 田口精一: バイオプラスチックのつくられ方とつくり方 蛋白質 核酸 酵素,50.3: 262-269 (2005) (in Japanese)
 "
"