Team:HokkaidoU Japan/Notebook/plastic Week 10
From 2012.igem.org
(→September 5th) |
(→Sequence=) |
||
| (24 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="hokkaidou-column-main"> | <div id="hokkaidou-column-main"> | ||
<!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 3rd== | + | ===September 3rd=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Single colony isolation== | + | ===Single colony isolation==== |
| - | + | ||
The colony of pGEM PhaCAB in JM109 was isolated to plate1 and plate2.<br/> | The colony of pGEM PhaCAB in JM109 was isolated to plate1 and plate2.<br/> | ||
| - | The colony of pGEM PhaCAB in | + | The colony of pGEM PhaCAB in DH5α was isolated to plate3.<br/> |
Plate 1~3 was started to incubate from 18:00. | Plate 1~3 was started to incubate from 18:00. | ||
| + | ====Colony PCR==== | ||
| + | We confirmed the length of constructs, [PhaB on pSB1C3] and [RBS-PhaC-RBS-PhaA on pSB1A2] by colony PCR.<br />The result showed ligation to take PhaB on pSB1C3 went well.<br />But ligation [RBS-PhaC on pSB1A2] with [RBS-PhaA] was failed. | ||
| - | + | ====Liquid culture==== | |
| - | + | We add 2 ml LB and 2 ul antibiotic to ideal colony suspension and begun to incubate at 37C, 180 rpm. | |
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==Liquid culture== | + | |
| - | + | ||
| - | We add | + | |
| - | + | ||
| - | + | ||
</div></div> | </div></div> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 4th== | + | ===September 4th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
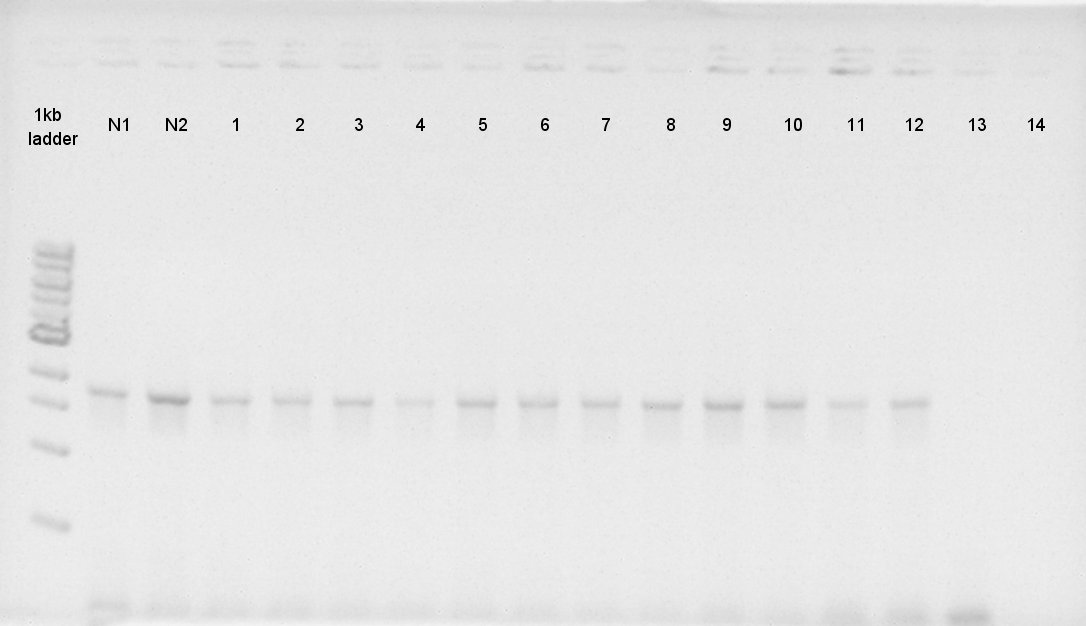
| + | [[image:HokkaidoU2012 120904RBSphaCcolop.jpg|thumb|Colony PCR result]] | ||
| + | ====Colony PCR==== | ||
| + | We confirmed the length of constructs, [RBS-PhaA on pSB1A2], [RBS-PhaC on pSB1A2] and [PhaB on pSB1C3] by colony PCR.<br />Ideal plasmids were liquid cultured and spreaded on LBplates. | ||
| + | ====PCR==== | ||
| + | PhaB on pSB1C3 dimer was multiplied by PCR.<br> | ||
| + | And then we got DNA solution of PhaB has prefix and suffix. | ||
</div></div> | </div></div> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 5th== | + | ===September 5th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| + | ====Plasmid extraction==== | ||
| + | We extracted the plasmids of [RBS-phaC] on pSB1A2 and [RBS-phaA] on pSB1A2 which was incubated from September 3rd. | ||
| + | Then we got 50 ul of DNA solutions. | ||
| - | === | + | ====Colony PCR==== |
| - | < | + | We confirmed the length of constructs, [RBS-PhaC-RBS-PhaA on pSB1A2] by colony PCR.<br />The result showed some of ligation [RBS-PhaC on pSB1A2] with [RBS-PhaA] went well. |
| - | Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit(NipponGenetics). | + | |
| + | ====Gel extraction==== | ||
| + | Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit (NipponGenetics). | ||
Got 50 ul of DNA solution of phaB. | Got 50 ul of DNA solution of phaB. | ||
| - | |||
| + | ====Liquid culture==== | ||
| + | The media of pGEM phaCAB was removed to polymer producing media. | ||
| + | </div></div> | ||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 6th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Plasmid extracton==== | ||
| + | The plasmids, [RBS-PhaC-RBS-PhaA] were extracted.<br/>And then we got 50ul DNA solution. | ||
| + | |||
| + | ====Sequence==== | ||
| + | We analyzed nucleotide sequence of construct made ever.<br /> | ||
| + | The result showed that PhaA sequence contained a mutation.<br />We decided to make construct including PhaA. | ||
</div></div> | </div></div> | ||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 7th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====harvesting==== | ||
| + | @Taguchi lab | ||
| + | The weight of new centrifuge tubes were measured.<br /> | ||
| + | |||
| + | JM109 | ||
| + | # pantothenic acid (+) 1.5 ml W:1.461 (W=tube weight with nothing inside.) | ||
| + | # pantothenic acid (-) 1.5 ml W:1.457 | ||
| + | # pantothenic acid (+) 0.7 ml W:1.576 | ||
| + | |||
| + | DH5a | ||
| + | # pantothenic acid (+) 1.5 ml W:1.461 | ||
| + | # pantothenic acid (-) 1.5 ml W:1.461 | ||
| + | # pantothenic acid (+) 0.7 ml W:1.460 | ||
| + | |||
| + | The cells were centrifuged and freeze dried for two days. | ||
| + | </div></div> | ||
<!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | ||
Latest revision as of 20:25, 26 September 2012
Contents |
September 3rd
Single colony isolation=
The colony of pGEM PhaCAB in JM109 was isolated to plate1 and plate2.
The colony of pGEM PhaCAB in DH5α was isolated to plate3.
Plate 1~3 was started to incubate from 18:00.
Colony PCR
We confirmed the length of constructs, [PhaB on pSB1C3] and [RBS-PhaC-RBS-PhaA on pSB1A2] by colony PCR.
The result showed ligation to take PhaB on pSB1C3 went well.
But ligation [RBS-PhaC on pSB1A2] with [RBS-PhaA] was failed.
Liquid culture
We add 2 ml LB and 2 ul antibiotic to ideal colony suspension and begun to incubate at 37C, 180 rpm.
September 4th
Colony PCR
We confirmed the length of constructs, [RBS-PhaA on pSB1A2], [RBS-PhaC on pSB1A2] and [PhaB on pSB1C3] by colony PCR.
Ideal plasmids were liquid cultured and spreaded on LBplates.
PCR
PhaB on pSB1C3 dimer was multiplied by PCR.
And then we got DNA solution of PhaB has prefix and suffix.
September 5th
Plasmid extraction
We extracted the plasmids of [RBS-phaC] on pSB1A2 and [RBS-phaA] on pSB1A2 which was incubated from September 3rd. Then we got 50 ul of DNA solutions.
Colony PCR
We confirmed the length of constructs, [RBS-PhaC-RBS-PhaA on pSB1A2] by colony PCR.
The result showed some of ligation [RBS-PhaC on pSB1A2] with [RBS-PhaA] went well.
Gel extraction
Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit (NipponGenetics). Got 50 ul of DNA solution of phaB.
Liquid culture
The media of pGEM phaCAB was removed to polymer producing media.
September 6th
Plasmid extracton
The plasmids, [RBS-PhaC-RBS-PhaA] were extracted.
And then we got 50ul DNA solution.
Sequence
We analyzed nucleotide sequence of construct made ever.
The result showed that PhaA sequence contained a mutation.
We decided to make construct including PhaA.
September 7th
harvesting
@Taguchi lab
The weight of new centrifuge tubes were measured.
JM109
- pantothenic acid (+) 1.5 ml W:1.461 (W=tube weight with nothing inside.)
- pantothenic acid (-) 1.5 ml W:1.457
- pantothenic acid (+) 0.7 ml W:1.576
DH5a
- pantothenic acid (+) 1.5 ml W:1.461
- pantothenic acid (-) 1.5 ml W:1.461
- pantothenic acid (+) 0.7 ml W:1.460
The cells were centrifuged and freeze dried for two days.
 "
"