Team:HokkaidoU Japan/Notebook/aggregation Week 8
From 2012.igem.org
| Line 228: | Line 228: | ||
[[image:|thumb|PCR result]] | [[image:|thumb|PCR result]] | ||
| - | + | </p> | |
==Aggregation check== | ==Aggregation check== | ||
| - | + | <p> | |
we cultured the E. coli, which transformed pBAD-RBS-Ag43-dT on pSB1AK3 in LBK. | we cultured the E. coli, which transformed pBAD-RBS-Ag43-dT on pSB1AK3 in LBK. | ||
We checked the construct by induction of L-arabinose after 16 hours incubate. | We checked the construct by induction of L-arabinose after 16 hours incubate. | ||
| Line 239: | Line 239: | ||
#Incubated at 37C 130rpm for 2hours and 30minutes. | #Incubated at 37C 130rpm for 2hours and 30minutes. | ||
#Placed tubes on the table at 30minutes. | #Placed tubes on the table at 30minutes. | ||
| + | </p> | ||
| + | </div><div> | ||
| - | |||
<!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | ||
</div> | </div> | ||
<br style="line-height: 0; clear: both;" /> | <br style="line-height: 0; clear: both;" /> | ||
{{Team:HokkaidoU_Japan/footer}} | {{Team:HokkaidoU_Japan/footer}} | ||
Revision as of 08:23, 21 August 2012
Contents |
August 20th
Single colony isolation
Single colony isolation of pBAD-RBS-Ag43-dT on pSB1AK3.
- Picked up one colony.
- Cultivation on LBK(dt,RBS,T7) and LBK(pLacI-RBS-Ag43) in hours.
Colony PCR
Colony PCR to confirm that whether the pT7 and RBS was successfully ligated with pSB1C3 or not.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(100bp up primer) | 0.5 ul |
| Reverse Primer(200bp down primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.2 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
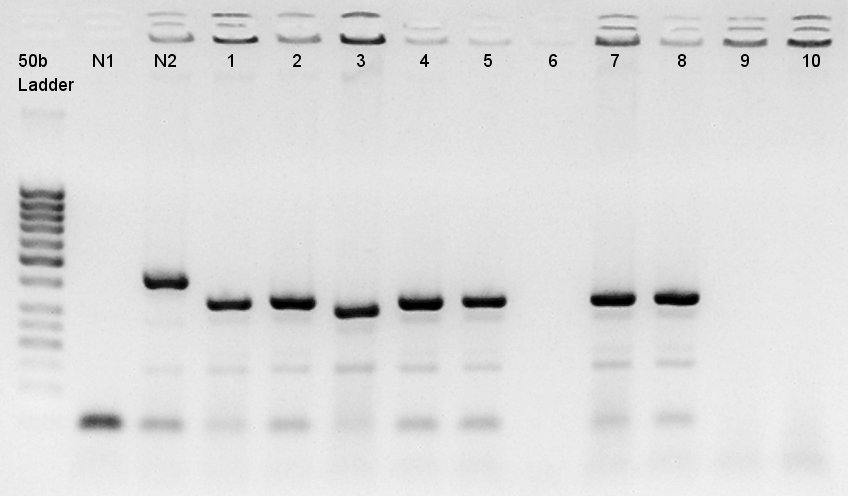
We used N1 (DW only) and N2 (pBAD(containing araC)-RBS on pSB1A3) as controls. Desired product is about 300~400bp.
The results showed that ligated DNA has 300 ~ 400 bp and the desired products would have 331bp if it were amplified by 100bp up primer and 200bp down primer. Thus we confirmed that pT7-RBS on pSB1C3 was successfully ligated without no.6,9,10 colonies, but these 3 solution were evapolated because of our mistake. We selected No.4 and 5 colony for liquid culture.
PCR
PCR of BBa_I13453 (pBAD only part, it is not contain araC,).
| DNA solution | 1 ul |
| KOD-Plus-NEO(Taq polymerase) | 1 ul |
| dNTP | 5 ul |
| MgSO4 | 3 ul |
| KOD-Plus-NEO Buffer | 5 ul |
| Forward Primer(Ag43-f4 primer: 10 uM) | 1 ul |
| Reverse Primer(PS-R primer: 10 uM) | 1 ul |
| DW | 33 ul |
| Total | 50 ul |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58 | 30 |
| 4 | 68 | 30 |
| 5 | 4 | HOLD |
Cycle:2~4 x 45
[[image:|thumb|PCR result]]
liquid culture
Liquid culture for 3 colonies of pBAD-RBS-Ag43-dT on pSB1AK3 (and pT7-RBS on pSB1C3 colony No.4 and 5 which were selected from the results of colony PCR).
- Added 2 ml of LBK(LBC) into culture tubes.
- Resuspended 1 colonies(Resuspended pre-cultivated 200ul of LB and colony solution).
- Incubated the tubes at 37C for 16 hours.
August 21th
PCR
PCR of pBAD(containing araC)-RBS. And, we checked the plasmid which we did mini-prep at August 18th is pBAD-RBS on pSB1A3 or not.
| DNA solution | 1 ul |
| KOD-Plus-NEO(Taq polymerase) | 1 ul |
| dNTP | 5 ul |
| MgSO4 | 3 ul |
| KOD-Plus-NEO Buffer | 5 ul |
| Forward Primer(Ag43-f4 primer: 10 uM) | 1 ul |
| Reverse Primer(PS-R primer: 10 uM) | 1 ul |
| DW | 33 ul |
| Total | 50 ul |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58 | 30 |
| 4 | 68 | 30 |
| 5 | 4 | HOLD |
Cycle:2~4 x 45
[[image:|thumb|PCR result]]
Aggregation check
we cultured the E. coli, which transformed pBAD-RBS-Ag43-dT on pSB1AK3 in LBK. We checked the construct by induction of L-arabinose after 16 hours incubate.
- 2 ml of liquid culture divided two culture. (made two 1 ml culture)
- Added 1 ml LBK in one culture as negative control.
- Added 900 ul LBK and 100 ul 20% L-arabinose.
- Incubated at 37C 130rpm for 2hours and 30minutes.
- Placed tubes on the table at 30minutes.
 "
"