Team:HokkaidoU Japan/Notebook/aggregation Week 5
From 2012.igem.org
(→PCR) |
|||
| (31 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="hokkaidou-column-main"> | <div id="hokkaidou-column-main"> | ||
<!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==July 30th== | + | ===July 30th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | == | + | ====Digestion==== |
| - | + | I tried to digest the plasmid extraction products again, and after Ethanol precipitation, we did electrophoresis. | |
| - | I tried to digest the | + | |
[[image:HokkaidoU2012_120730_ag43-f1-e-s.jpg|thumb|digestion result]] | [[image:HokkaidoU2012_120730_ag43-f1-e-s.jpg|thumb|digestion result]] | ||
| - | + | ====Transformation==== | |
| - | == | + | I think that the E. coli which we used transformation is BL21(DE3)pLysS. |
| - | + | So, the E. coli could grow on LBC plate.<br /> | |
| - | I think that the E. coli which we | + | I'm going to do transformation into DH5α. |
| - | So, the E. coli could | + | |
| - | I'm going to do transformation | + | |
| - | Transformation | + | Transformation plasmid DNA ligated at 25th (pT7-RBS-Ag43-dT on pSB1K3) into DH5α. |
#Added 2 ul of plasmid DNA to 50 ul of thawed competent cells on ice. | #Added 2 ul of plasmid DNA to 50 ul of thawed competent cells on ice. | ||
#Incubated on ice for 30 min. | #Incubated on ice for 30 min. | ||
| - | #Added 200 ul of LB then incubated the cells for 2 | + | #Added 200 ul of LB then incubated the cells for 2 hrs at 37C. |
#Prepared and Labeled two petri dishes with LBK. | #Prepared and Labeled two petri dishes with LBK. | ||
#Plate 200 ul of the transformation onto first dish and spread. | #Plate 200 ul of the transformation onto first dish and spread. | ||
| - | #Added 450 ul of LB to 50 ul of the transformation and | + | #Added 450 ul of LB to 50 ul of the transformation and spread 200 ul of it onto second dish. |
| - | #Incubated the plates at 37C for 14 | + | #Incubated the plates at 37C for 14 hrs. |
| - | </ | + | <br style="line-height: 0; clear: both;" /> |
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==July | + | ===July 31st=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | == | + | ====Liquid culture==== |
| - | + | ||
Liquid culture for pT7-RBS-Ag43-dT on pSB1K3. | Liquid culture for pT7-RBS-Ag43-dT on pSB1K3. | ||
#Added 2 ml of LBK into culture tubes. | #Added 2 ml of LBK into culture tubes. | ||
#Resuspended colonies. | #Resuspended colonies. | ||
| - | #Incubated the tubes at 37C for | + | #Incubated the tubes at 37C for 18 hrs. |
| - | + | ||
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 1st== | + | ===August 1st=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | == | + | ====Plasmid extraction==== |
| - | + | Plasmid extraction of pT7-RBS-Ag43-dT which had been incubated from 31st July. We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions. | |
| - | + | [[image:HokkaidoU2012 120801 pt7-rbs-ag43-dt.jpg|thumb|Plasmid extraction result]] | |
| - | [[image:HokkaidoU2012 120801 pt7-rbs-ag43-dt.jpg|thumb| | + | |
| - | + | ||
| - | ==Digestion== | + | ====Digestion==== |
| - | + | '''pT7-RBS(20 ng/ul) '''=No. 9 | |
| - | '''pT7-RBS(20 ng/ul) '''= | + | <br /> |
| - | + | SpeI using 10xH | |
| - | SpeI 10xH | + | |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 78: | Line 69: | ||
|} | |} | ||
| - | SpeI 10xM | + | |
| + | SpeI using 10xM | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 98: | Line 90: | ||
| - | '''pT7-RBS(30 ng/ul) '''= | + | '''pT7-RBS(30 ng/ul) '''=No. 10 |
| - | + | <br /> | |
| - | SpeI 10xH | + | SpeI using 10xH |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 119: | Line 111: | ||
|} | |} | ||
| - | SpeI 10xM | + | SpeI using 10xM |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 140: | Line 132: | ||
[[image:HokkaidoU2012 120801 pt7-rbs-digS.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120801 pt7-rbs-digS.jpg|thumb|digestion result]] | ||
| - | We couldn't | + | We couldn't digested them exactly, so we tried to digest once more time.<br /> |
| - | '''pT7-RBS(20 ng/ul) '''= | + | '''pT7-RBS(20 ng/ul) '''=No. 9<br /> |
SpeI 10xH | SpeI 10xH | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 161: | Line 153: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | + | ||
| - | + | ====Liquid culture==== | |
| - | ==Liquid culture== | + | |
| - | + | ||
Liquid culture for pBAD-RBS on pSB1K3. | Liquid culture for pBAD-RBS on pSB1K3. | ||
#Added 2 ml of LBK into culture tube. | #Added 2 ml of LBK into culture tube. | ||
#Scraped the surface of glycerol stock of construct. | #Scraped the surface of glycerol stock of construct. | ||
#Incubated the tube at 33C. | #Incubated the tube at 33C. | ||
| - | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 2nd== | + | ===August 2nd=== |
| - | <div | + | <div class="hokkaidou-section"> |
| - | + | ====Preparing chemical competent cell==== | |
| - | ==Preparing chemical competent cell== | + | Preparing chemical competent cell of BL21, JM109 and DH5α. |
| - | Preparing chemical competent cell of BL21, JM109 and | + | |
Chemical competent cell made in each E. coli strains. | Chemical competent cell made in each E. coli strains. | ||
| - | + | <br /><br /> | |
Our competent cell Protocol | Our competent cell Protocol | ||
| - | # | + | #Did single colony isolation on LB plate. |
| - | # | + | #Incubated the plate for 15-19 hours at 37C. |
| - | # | + | #Lifted colony of E. coli into 2 ml LB. |
| - | # | + | #Incubated at 37C for 12-16 hrs at 180-200 rpm. |
| - | # | + | #Transfered 30 ul, 100 ul, 300 ul of the culture into 100 ml of LB. |
| - | # | + | #Incubated cells at 20C (over 24 hrs) at 140 rpm. |
| - | # | + | #Selected culture by measuring OD 600. |
| - | # | + | #Incubated the 300 ml flask for 10 min on ice. |
| - | # | + | #Transfered the culture into two 50 ml Falcon tubes. |
| - | # | + | #Centrifuged at 3000 rpm, 4C for 20 min (TOMY LX-120 rotor), and discard supernatant. |
| - | # | + | #Suspended the pellet in ice-cold 15 ml of TB (Transformation Buffer).(7.5 ml/tube) |
| - | # | + | #Collected them to one tube. |
| - | # | + | #Centrifuged at 3000 rpm, 4C for 20 min (TOMY LX-120 rotor), and discard supernatant. |
| - | # | + | #Suspended the pellet in ice-cold 3.2 ml of TB. |
| - | # | + | #Instilled 0.24 ml of DMSO in precipitant. |
| - | # | + | #Incubated the 50 ml Falcon tube for 10 min on ice. |
| - | # | + | #Divided 50 ul of solutions in each 0.5 ml tubes. |
| - | #Freezed the suspension | + | #Freezed the suspension by liquid nitrogen. |
| - | # | + | #Stored at â80C. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ====Electrophoresis==== | ||
| + | Electrophoresis of digestion result of August 1st.(pT7-RBS on pSB1K3 digested by SpeI) | ||
[[image:HokkaidoU2012 120802 pT7-RBS on pSB1K3 SpeI.jpg|thumb|Electrophoresis result]] | [[image:HokkaidoU2012 120802 pT7-RBS on pSB1K3 SpeI.jpg|thumb|Electrophoresis result]] | ||
| - | There were low concentration band | + | There were low concentration band above thick band. This thick band was same as digestion minus band. |
| - | + | ||
| - | ==Digestion== | + | ====Digestion==== |
| - | + | Digestion of pT7-RBS on pSB1K3 (more fresh one) with SpeI. | |
| - | Digestion of pT7-RBS on pSB1K3(more fresh one) | + | |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 234: | Line 215: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 3rd== | + | ===August 3rd=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Electrophoresis== | + | ====Electrophoresis==== |
| - | + | Electrophoresis for the result of digestion. | |
| - | Electrophoresis for the result of digestion | + | |
[[image:HokkaidoU2012 120803 pt7-rbs-dspe1.jpg|thumb|Electrophoresis result]] | [[image:HokkaidoU2012 120803 pt7-rbs-dspe1.jpg|thumb|Electrophoresis result]] | ||
| - | |||
| - | ==Gel extraction== | + | ====Gel extraction==== |
| - | + | ||
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | ||
| - | |||
| - | ==Ethanol precipitation== | + | ====Ethanol precipitation==== |
| - | + | ||
Ethanol precipitation for digestion and gel extraction product. | Ethanol precipitation for digestion and gel extraction product. | ||
| - | #Added | + | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125ul of 100% ethanol. |
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 10 min at 4C. |
| - | # | + | #Removed supernatant and added 220ul of 70% ethanol. |
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 5 min at 4C. |
| - | # | + | #Removed supernatant and dried out at room temperature after that added 10 ul of DW. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ====Ligation==== | ||
| + | Ligation of pT7-RBS on pSB1K3 (digested Spe1) as vector and Ag43-dT (digested Spe1 and Xba1 and HindIII) as insert. | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
| Line 300: | Line 273: | ||
|Hold | |Hold | ||
|} | |} | ||
| - | |||
| - | ==Transformation== | + | |
| - | + | ====Transformation==== | |
| - | Transformation | + | Transformation plasmid DNA ligated product (pT7-RBS-Ag43-dT on pSB1K3) into DH5α. |
| - | #Added | + | #Added 1 ul of plasmid DNA to 50 ul of thawed competent cells on ice. |
#Incubated on ice for 30min. | #Incubated on ice for 30min. | ||
| - | #Added | + | #Added 600 ul of LB then incubated the cells for 2 hrs at 37C. |
#Prepared and Labeled two petri dishes with LBK. | #Prepared and Labeled two petri dishes with LBK. | ||
| - | # | + | #Spread 300 ul of the transformation onto first dish. |
| - | #Added | + | #Added 900 ul of LB to 100 ul of the transformation and spread 300 ul of it onto second dish. |
| - | #Incubated the plates at 37C for 15 hrs 45 min | + | #Incubated the plates at 37C for 15 hrs 45 min. |
| - | + | ||
| - | ==PCR== | + | ====PCR==== |
| - | + | [[image:HokkaidoU2012 120803 Ag43 PCR(Forward=ag43-f4 Reverse=PS-R).jpg|thumb|PCR result]] | |
PCR to confirm Ag43-f4 primer. | PCR to confirm Ag43-f4 primer. | ||
| - | |||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | | | + | |1 ul |
|- | |- | ||
|KOD-Plus-NEO(Taq polymerase) | |KOD-Plus-NEO(Taq polymerase) | ||
| - | | | + | |1 ul |
|- | |- | ||
|dNTP | |dNTP | ||
| - | | | + | |5 ul |
|- | |- | ||
|MgSO4 | |MgSO4 | ||
| - | | | + | |3 ul |
|- | |- | ||
|KOD-Plus-NEO Buffer | |KOD-Plus-NEO Buffer | ||
| - | | | + | |5 ul |
|- | |- | ||
| - | | | + | |Ag43-f4 primer |
| - | | | + | |1 ul |
|- | |- | ||
| - | | | + | |PS-R primer |
| - | | | + | |1 ul |
|- | |- | ||
|DW | |DW | ||
| - | | | + | |33 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |50 ul |
|} | |} | ||
| Line 377: | Line 347: | ||
Cycle:2~4 x 45 | Cycle:2~4 x 45 | ||
| + | From this result, we could see that the ag43-f4 primer had worked and DNA of last 500~600 bp of ag43 to BioBrick suffix were increased. | ||
| - | + | <br style="line-height: 0; clear: both;" /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div></div> | </div></div> | ||
| + | <div class="hokkaidou-notebook-daily"> | ||
| - | <div class="hokkaidou- | + | ===August 4th=== |
| - | == | + | <div class="hokkaidou-section"> |
| - | + | ====Colony PCR==== | |
| - | ==Colony PCR== | + | |
| - | + | ||
Colony PCR to confirm that whether the Ag43-dT was successfully ligated with pT7-RBS on pSB1K3 or not. | Colony PCR to confirm that whether the Ag43-dT was successfully ligated with pT7-RBS on pSB1K3 or not. | ||
| - | |||
| - | |||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 452: | Line 414: | ||
[[image:HokkaidoU2012 120804 pt7-rbs-ag43-dt coloP.jpg|thumb|Colony PCR result]] | [[image:HokkaidoU2012 120804 pt7-rbs-ag43-dt coloP.jpg|thumb|Colony PCR result]] | ||
| - | We thought that No.5 and 9 | + | We thought that colonies No. 5 and 9 were exactly transformed into E. coli. and colony of No. 10 had high concentration band. |
| - | Next step, we resuspended these three colonies and | + | Next step, we resuspended these three colonies and incubated. |
| - | + | ||
| - | ==Liquid culture== | + | ====Liquid culture==== |
| - | + | ||
Liquid culture of colonies passed the colony PCR test. | Liquid culture of colonies passed the colony PCR test. | ||
#Added 2 ml of LBK into culture tubes. | #Added 2 ml of LBK into culture tubes. | ||
#Resuspended 3 colonies. | #Resuspended 3 colonies. | ||
#Incubated the tubes at 37C for 13 hrs. | #Incubated the tubes at 37C for 13 hrs. | ||
| - | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 5th== | + | ===August 5th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | == | + | ====Plasmid extraction==== |
| - | + | Plasmid extraction of incubated medium from yesterday. We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions. | |
| - | + | [[image:HokkaidoU2012 120805 pt7-rbs-ag43-dt.jpg|thumb|Plasmid extraction results]] | |
| - | + | ||
| - | [[image:HokkaidoU2012 120805 pt7-rbs-ag43-dt.jpg|thumb| | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ====Digestion==== | ||
| + | Digested pT7-RBS on pSB1K3 and pT7-RBS (once digestioned with SpeI) with speI.<br /> | ||
pT7-RBS on pSB1K3 | pT7-RBS on pSB1K3 | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 520: | Line 473: | ||
|10 ul | |10 ul | ||
|} | |} | ||
| + | |||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 541: | Line 495: | ||
[[image:HokkaidoU2012 120805 pT7-RBS digestion SpeI (no.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120805 pT7-RBS digestion SpeI (no.jpg|thumb|digestion result]] | ||
| - | + | From this result, the bottom band was digested incorrectly and middle band was digested correctly, we thought. Was the above band something contaminated in the DNA solution? | |
| - | + | ====Gel extraction==== | |
| - | ==Gel extraction== | + | Gel extraction of middle band. We thought this band would be the exactly digested DNA fragment. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. |
| - | + | ||
| - | Gel extraction of middle band | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ====Sequencing==== | ||
| + | Sequencing to confirm what kind of DNA we made. | ||
{| | {| | ||
| Line 557: | Line 507: | ||
|primer | |primer | ||
|- | |- | ||
| - | |Ag43 | + | |Ag43 plasmid extraction product |
|100bp-up forward primer, | |100bp-up forward primer, | ||
|ag43-f1, | |ag43-f1, | ||
| Line 600: | Line 550: | ||
|- | |- | ||
|template DNA | |template DNA | ||
| - | | | + | |1 ul |
|- | |- | ||
|Ready Reaction Premix | |Ready Reaction Premix | ||
| - | | | + | |1 ul |
|- | |- | ||
|5x Sequencing Buffer | |5x Sequencing Buffer | ||
| - | |1. | + | |1.5 ul |
|- | |- | ||
|H2O | |H2O | ||
| - | | | + | |5 ul |
|- | |- | ||
| - | |Primer( | + | |Primer(1 pmol/ul) |
| - | |1. | + | |1.5 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |10 ul |
|} | |} | ||
| Line 644: | Line 594: | ||
| - | To | + | To extract the PCR product, we did ethanol precipitation. |
| + | <br /> | ||
| + | Ethanol precipitation for sequencing. | ||
| + | #Added 10 ul of H2O, 2 ul of NaoAc, 1 ul of glycogen and 50 ul of 100% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 15 min at 26C. | ||
| + | #Removed supernatant and added 100ul of 70% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 5 min at 26C. | ||
| + | #Removed supernatant and dried out at room temperature after that added 10 ul of Hi-Di. | ||
| + | Then ran a sequencing machine.(ByoDye Terminator v3.1 Cycle Sequencing Kit)<br /> | ||
| + | We could not get the sequencing data. | ||
| - | + | ====Digestion==== | |
| - | + | Digesting of Ag43-dT (digested by SpeI and XbaI) with HindIII. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==Digestion== | + | |
| - | + | ||
| - | + | ||
| - | + | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| Line 680: | Line 625: | ||
|20 ul | |20 ul | ||
|} | |} | ||
| - | |||
[[image:HokkaidoU2012 120805 Ag43-dT on pSB1AK3(d+ with XbaI & SpeI) digestion with HindIII.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120805 Ag43-dT on pSB1AK3(d+ with XbaI & SpeI) digestion with HindIII.jpg|thumb|digestion result]] | ||
| + | From this result, we confirmed that the pSB1AK3 was successfully digested by HindIII. | ||
| - | + | ====Gel extraction==== | |
| - | + | ||
| - | + | ||
| - | ==Gel extraction== | + | |
| - | + | ||
Gel extraction for digestion production. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | Gel extraction for digestion production. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| + | |||
<!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | ||
</div> | </div> | ||
<br style="line-height: 0; clear: both;" /> | <br style="line-height: 0; clear: both;" /> | ||
{{Team:HokkaidoU_Japan/footer}} | {{Team:HokkaidoU_Japan/footer}} | ||
Latest revision as of 03:32, 27 September 2012
Contents |
July 30th
Digestion
I tried to digest the plasmid extraction products again, and after Ethanol precipitation, we did electrophoresis.
Transformation
I think that the E. coli which we used transformation is BL21(DE3)pLysS.
So, the E. coli could grow on LBC plate.
I'm going to do transformation into DH5α.
Transformation plasmid DNA ligated at 25th (pT7-RBS-Ag43-dT on pSB1K3) into DH5α.
- Added 2 ul of plasmid DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Added 200 ul of LB then incubated the cells for 2 hrs at 37C.
- Prepared and Labeled two petri dishes with LBK.
- Plate 200 ul of the transformation onto first dish and spread.
- Added 450 ul of LB to 50 ul of the transformation and spread 200 ul of it onto second dish.
- Incubated the plates at 37C for 14 hrs.
July 31st
Liquid culture
Liquid culture for pT7-RBS-Ag43-dT on pSB1K3.
- Added 2 ml of LBK into culture tubes.
- Resuspended colonies.
- Incubated the tubes at 37C for 18 hrs.
August 1st
Plasmid extraction
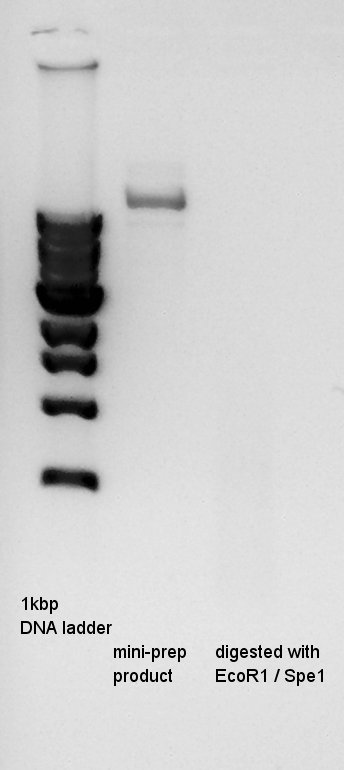
Plasmid extraction of pT7-RBS-Ag43-dT which had been incubated from 31st July. We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions.
Digestion
pT7-RBS(20 ng/ul) =No. 9
SpeI using 10xH
| DNA solution | 4.5 ul |
| SpeI | 1 ul |
| 10xH buffer | 1 ul |
| DW | 3.5 ul |
| Total | 10 ul |
SpeI using 10xM
| DNA solution | 4.5 ul |
| SpeI | 1 ul |
| 10xM buffer | 1 ul |
| DW | 3.5 ul |
| Total | 10 ul |
pT7-RBS(30 ng/ul) =No. 10
SpeI using 10xH
| DNA solution | 3 ul |
| SpeI | 1 ul |
| 10xH buffer | 1 ul |
| DW | 5 ul |
| Total | 10 ul |
SpeI using 10xM
| DNA solution | 3 ul |
| SpeI | 1 ul |
| 10xM buffer | 1 ul |
| DW | 5 ul |
| Total | 10 ul |
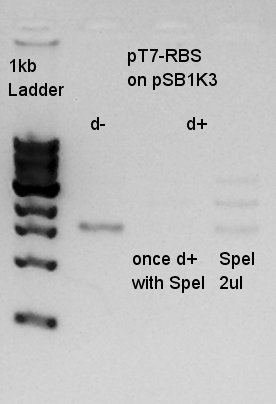
We couldn't digested them exactly, so we tried to digest once more time.
pT7-RBS(20 ng/ul) =No. 9
SpeI 10xH
| DNA solution | 4.5 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 12.5 ul |
| Total | 20 ul |
Liquid culture
Liquid culture for pBAD-RBS on pSB1K3.
- Added 2 ml of LBK into culture tube.
- Scraped the surface of glycerol stock of construct.
- Incubated the tube at 33C.
August 2nd
Preparing chemical competent cell
Preparing chemical competent cell of BL21, JM109 and DH5α.
Chemical competent cell made in each E. coli strains.
Our competent cell Protocol
- Did single colony isolation on LB plate.
- Incubated the plate for 15-19 hours at 37C.
- Lifted colony of E. coli into 2 ml LB.
- Incubated at 37C for 12-16 hrs at 180-200 rpm.
- Transfered 30 ul, 100 ul, 300 ul of the culture into 100 ml of LB.
- Incubated cells at 20C (over 24 hrs) at 140 rpm.
- Selected culture by measuring OD 600.
- Incubated the 300 ml flask for 10 min on ice.
- Transfered the culture into two 50 ml Falcon tubes.
- Centrifuged at 3000 rpm, 4C for 20 min (TOMY LX-120 rotor), and discard supernatant.
- Suspended the pellet in ice-cold 15 ml of TB (Transformation Buffer).(7.5 ml/tube)
- Collected them to one tube.
- Centrifuged at 3000 rpm, 4C for 20 min (TOMY LX-120 rotor), and discard supernatant.
- Suspended the pellet in ice-cold 3.2 ml of TB.
- Instilled 0.24 ml of DMSO in precipitant.
- Incubated the 50 ml Falcon tube for 10 min on ice.
- Divided 50 ul of solutions in each 0.5 ml tubes.
- Freezed the suspension by liquid nitrogen.
- Stored at â80C.
Electrophoresis
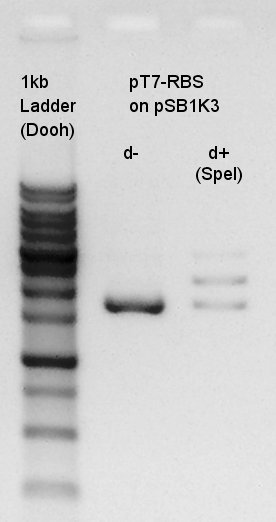
Electrophoresis of digestion result of August 1st.(pT7-RBS on pSB1K3 digested by SpeI)
There were low concentration band above thick band. This thick band was same as digestion minus band.
Digestion
Digestion of pT7-RBS on pSB1K3 (more fresh one) with SpeI.
| DNA solution | 4 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 13 ul |
| Total | 20 ul |
August 3rd
Electrophoresis
Electrophoresis for the result of digestion.
Gel extraction
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
Ethanol precipitation
Ethanol precipitation for digestion and gel extraction product.
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125ul of 100% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and added 220ul of 70% ethanol.
- Centrifuged at 15000 rpm, 5 min at 4C.
- Removed supernatant and dried out at room temperature after that added 10 ul of DW.
Ligation
Ligation of pT7-RBS on pSB1K3 (digested Spe1) as vector and Ag43-dT (digested Spe1 and Xba1 and HindIII) as insert.
| pT7-RBS | 2 ul |
| Ag43-dT | 4 ul |
| Ligation Mighty Mix(TAKARA) | 6 ul |
| Total | 12 ul |
Ligation reaction time was written below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation
Transformation plasmid DNA ligated product (pT7-RBS-Ag43-dT on pSB1K3) into DH5α.
- Added 1 ul of plasmid DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30min.
- Added 600 ul of LB then incubated the cells for 2 hrs at 37C.
- Prepared and Labeled two petri dishes with LBK.
- Spread 300 ul of the transformation onto first dish.
- Added 900 ul of LB to 100 ul of the transformation and spread 300 ul of it onto second dish.
- Incubated the plates at 37C for 15 hrs 45 min.
PCR
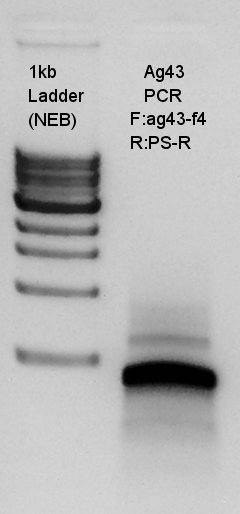
PCR to confirm Ag43-f4 primer.
| DNA solution | 1 ul |
| KOD-Plus-NEO(Taq polymerase) | 1 ul |
| dNTP | 5 ul |
| MgSO4 | 3 ul |
| KOD-Plus-NEO Buffer | 5 ul |
| Ag43-f4 primer | 1 ul |
| PS-R primer | 1 ul |
| DW | 33 ul |
| Total | 50 ul |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58 | 30 |
| 4 | 68 | 60 |
| 5 | 4 | HOLD |
Cycle:2~4 x 45
From this result, we could see that the ag43-f4 primer had worked and DNA of last 500~600 bp of ag43 to BioBrick suffix were increased.
August 4th
Colony PCR
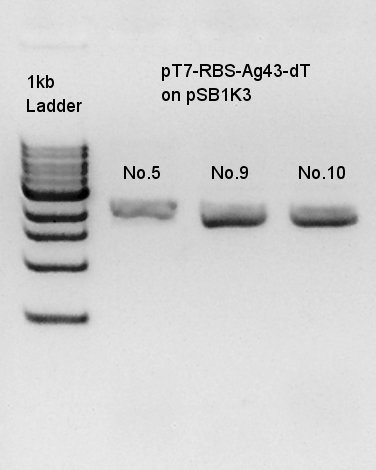
Colony PCR to confirm that whether the Ag43-dT was successfully ligated with pT7-RBS on pSB1K3 or not.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(Ag43-f4 primer) | 0.5 ul |
| Reverse Primer(PS-R primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 58 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2 (Ag43-dT on pSB1AK3) as controls. Desired product is about 500~600bp.
We thought that colonies No. 5 and 9 were exactly transformed into E. coli. and colony of No. 10 had high concentration band. Next step, we resuspended these three colonies and incubated.
Liquid culture
Liquid culture of colonies passed the colony PCR test.
- Added 2 ml of LBK into culture tubes.
- Resuspended 3 colonies.
- Incubated the tubes at 37C for 13 hrs.
August 5th
Plasmid extraction
Plasmid extraction of incubated medium from yesterday. We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions.
Digestion
Digested pT7-RBS on pSB1K3 and pT7-RBS (once digestioned with SpeI) with speI.
pT7-RBS on pSB1K3
| DNA solution | 8 ul |
| SpeI | 2 ul |
| 10xM buffer | 2 ul |
| DW | 8 ul |
| Total | 20 ul |
pT7-RBS (once digestioned with SpeI)
| DNA solution | 4 ul |
| SpeI | 1 ul |
| 10xM buffer | 1 ul |
| DW | 4 ul |
| Total | 10 ul |
| DNA solution | 4 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 13 ul |
| Total | 20 ul |
From this result, the bottom band was digested incorrectly and middle band was digested correctly, we thought. Was the above band something contaminated in the DNA solution?
Gel extraction
Gel extraction of middle band. We thought this band would be the exactly digested DNA fragment. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
Sequencing
Sequencing to confirm what kind of DNA we made.
| DNA | primer | ||||
| Ag43 plasmid extraction product | 100bp-up forward primer, | ag43-f1, | ag43-f2, | ag43-f3, | ag43-f4 |
| K542009 | 100bp-up forward primer, | ag43-f1, | ag43-f2, | ag43-f3, | ag43-f4 |
| pT7-RBS on pSB1K3 | 100bp-up forward primer | ||||
| pT7-RBS on pSB1C3 | 100bp-up forward primer | ||||
| Ag43-dT on pSB1AK3 | 100bp-up forward primer, | ag43-f1, | ag43-f2, | ag43-f3, | ag43-f4 |
| Ag43-dT on pSB1T3 | 100bp-up forward primer, | ag43-f1, | ag43-f2, | ag43-f3, | ag43-f4 |
| pT7-RBS on pSB1K3 | 100bp-up forward primer |
Sequencing PCR
| template DNA | 1 ul |
| Ready Reaction Premix | 1 ul |
| 5x Sequencing Buffer | 1.5 ul |
| H2O | 5 ul |
| Primer(1 pmol/ul) | 1.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 96 | 10 |
| 2 | 50 | 5 |
| 3 | 60 | 240 |
| 4 | 4 | HOLD |
Cycle:1~3 x 25
To extract the PCR product, we did ethanol precipitation.
Ethanol precipitation for sequencing.
- Added 10 ul of H2O, 2 ul of NaoAc, 1 ul of glycogen and 50 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 26C.
- Removed supernatant and added 100ul of 70% ethanol.
- Centrifuged at 15000 rpm, 5 min at 26C.
- Removed supernatant and dried out at room temperature after that added 10 ul of Hi-Di.
Then ran a sequencing machine.(ByoDye Terminator v3.1 Cycle Sequencing Kit)
We could not get the sequencing data.
Digestion
Digesting of Ag43-dT (digested by SpeI and XbaI) with HindIII.
| DNA solution | 10 ul |
| HindIII | 1 ul |
| 10xM buffer | 2 ul |
| DW | 7 ul |
| Total | 20 ul |
From this result, we confirmed that the pSB1AK3 was successfully digested by HindIII.
Gel extraction
Gel extraction for digestion production. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
 "
"