Team:HokkaidoU Japan/Notebook/aggregation Week 1
From 2012.igem.org
| Line 25: | Line 25: | ||
<p> | <p> | ||
Transformation of BBa_B0015(dT), B0034(RBS), I179005(pT7), K346007(Ag43), and K542009(pLacI-RBS-Ag43) in DH5α | Transformation of BBa_B0015(dT), B0034(RBS), I179005(pT7), K346007(Ag43), and K542009(pLacI-RBS-Ag43) in DH5α | ||
| - | #Added each DNA solutions ( | + | #Added each DNA solutions (1 ul) into DH5α comeptent cell and standed on ice in 30 min. |
| - | #Cultivated on LBA(dt,RBS,T7) and LBC(Ag43, pLacI-RBS-Ag43). E.coli cultivate in LBC was pre-cultivated in 2 hrs. pT7 was | + | #Cultivated on LBA(dt,RBS,T7) and LBC(Ag43, pLacI-RBS-Ag43). E.coli cultivate in LBC was pre-cultivated in 2 hrs. pT7 was 21 hrs and Others were 20 hrs cultivated. |
</p> | </p> | ||
</div></div> | </div></div> | ||
| Line 41: | Line 41: | ||
Transformation of K346007(Ag43) in DH5α. | Transformation of K346007(Ag43) in DH5α. | ||
| - | #Add DNA solution( | + | #Add DNA solution(1 ul) into DH5α comeptent cell and stand on ice in 30 min. |
| - | #Pre-cultivated in | + | #Pre-cultivated in 2 hrs. |
#Cultivated on LBC in 21hrs. | #Cultivated on LBC in 21hrs. | ||
</p> | </p> | ||
| Line 49: | Line 49: | ||
Single colony isolation of BBa_B0015, B0034, I179005 and K542009. | Single colony isolation of BBa_B0015, B0034, I179005 and K542009. | ||
#Picked up one colony. | #Picked up one colony. | ||
| - | #Cultivation on LBA(dt,RBS,T7) and LBC(pLacI-RBS-Ag43) in | + | #Cultivation on LBA(dt,RBS,T7) and LBC(pLacI-RBS-Ag43) in 14 hrs and 30 mins |
'''BBa_K542009 was Ag43 only part! And the part didn't have Biobrick suffix.''' | '''BBa_K542009 was Ag43 only part! And the part didn't have Biobrick suffix.''' | ||
</p> | </p> | ||
| Line 63: | Line 63: | ||
Liquid culture in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43) | Liquid culture in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43) | ||
#Picked up two colonies from each plates. | #Picked up two colonies from each plates. | ||
| - | #One colony was resuspended in | + | #One colony was resuspended in 1 ml LB (A or C), and other colony was resuspended in 2 ml LB(A or C). 1 ml is for glycerol stocks and 2 ml is for mini-prep. |
| - | # | + | #16 hrs Cultivation. |
</p> | </p> | ||
===Single colony isolation=== | ===Single colony isolation=== | ||
| Line 78: | Line 78: | ||
<div> | <div> | ||
===Liquid culture=== | ===Liquid culture=== | ||
| + | <p> | ||
Liquid culture in LBC(Ag43). | Liquid culture in LBC(Ag43). | ||
#Resuspended two colonies from each plates. | #Resuspended two colonies from each plates. | ||
| - | #Both of colonies were dipped in | + | #Both of colonies were dipped in 2 ml LBC, and then we cultivate them in 38C.<br> |
'''However, one of them cultivated only 8 hours. It's for glycerol stock.''' | '''However, one of them cultivated only 8 hours. It's for glycerol stock.''' | ||
| + | </p> | ||
| + | |||
===3A assembly=== | ===3A assembly=== | ||
| + | <p> | ||
Assembled pT7, RBS and pSB1C3 by 3A assembly. | Assembled pT7, RBS and pSB1C3 by 3A assembly. | ||
This 3A assembly is our first try! | This 3A assembly is our first try! | ||
| + | </p> | ||
| + | |||
===mini-prep=== | ===mini-prep=== | ||
| + | <p> | ||
#mini-prep of dT,RBS,pT7 and pLacI-RBS-Ag43. We used FastGene Plasmid Mini Kit(Nippon Gene) | #mini-prep of dT,RBS,pT7 and pLacI-RBS-Ag43. We used FastGene Plasmid Mini Kit(Nippon Gene) | ||
| - | #Got | + | #Got 50 ul of DNA solutions. |
| + | </p> | ||
| + | |||
===Glycerol stock=== | ===Glycerol stock=== | ||
| + | <p> | ||
glycerol stocks of dT,RBS,pT7 and pLacI-RBS-Ag43. | glycerol stocks of dT,RBS,pT7 and pLacI-RBS-Ag43. | ||
| - | #Parts written above were cultivated in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43) | + | #Parts written above were cultivated in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43) 1 ml in 16 hrs 30 min. |
#Add glycerol and Freeze at -80C | #Add glycerol and Freeze at -80C | ||
| + | </p> | ||
| + | |||
===Electrophoresis=== | ===Electrophoresis=== | ||
| + | <p> | ||
[[Image:120707_I719005_B0034_B0015_K542009_mini-prep_umeuchi.jpg|thumb|Erectrophoresis result]] | [[Image:120707_I719005_B0034_B0015_K542009_mini-prep_umeuchi.jpg|thumb|Erectrophoresis result]] | ||
Electrophoresis to predict concentration of mini-prep products(dT,RBS,pT7 and pLacI-RBS-Ag43). | Electrophoresis to predict concentration of mini-prep products(dT,RBS,pT7 and pLacI-RBS-Ag43). | ||
#Used 1% agarose gel. | #Used 1% agarose gel. | ||
#Added EtBr into TBE buffer and started Pre-migration. | #Added EtBr into TBE buffer and started Pre-migration. | ||
| - | #Migrated '''1. | + | #Migrated '''1.2 ul of DNA solutions (1 ul is mini-prep products and 0.2 ul is Loading Dye)''' in 35 min. |
#Took a photograph of 1% agarose gel that finished electrophoresis. | #Took a photograph of 1% agarose gel that finished electrophoresis. | ||
| + | </p> | ||
| + | |||
===Digestion=== | ===Digestion=== | ||
| + | <p> | ||
Digestion of I719005, B0034 and pSB1K3 | Digestion of I719005, B0034 and pSB1K3 | ||
| Line 105: | Line 121: | ||
All parts were reacted in 30ul digestion mix. | All parts were reacted in 30ul digestion mix. | ||
| - | *I719005( | + | *I719005(40 ng/ul) |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | |12. | + | |12.5 ul |
|- | |- | ||
|EcoRI | |EcoRI | ||
| - | | | + | |1 ul |
|- | |- | ||
|SpeI | |SpeI | ||
| - | | | + | |1 ul |
|- | |- | ||
|10xH Buffer | |10xH Buffer | ||
| - | | | + | |3 ul |
|- | |- | ||
|DW | |DW | ||
| - | |12. | + | |12.5 ul |
|} | |} | ||
| - | *B0034( | + | *B0034(40 ng/ul) |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | |12. | + | |12.5 ul |
|- | |- | ||
|XbaI | |XbaI | ||
| - | | | + | |1 ul |
|- | |- | ||
|PstI | |PstI | ||
| - | | | + | |1 ul |
|- | |- | ||
|10xM Buffer | |10xM Buffer | ||
| - | | | + | |3 ul |
|- | |- | ||
|DW | |DW | ||
| - | |12. | + | |12.5 ul |
|} | |} | ||
| - | *pSB1K3( | + | *pSB1K3(25 ng/ul) |
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | | | + | |12 ul |
|- | |- | ||
|EcoRI | |EcoRI | ||
| - | | | + | |1 ul |
|- | |- | ||
|PstI | |PstI | ||
| - | | | + | |1 ul |
|- | |- | ||
|10xH Buffer | |10xH Buffer | ||
| - | | | + | |3 ul |
|- | |- | ||
|DW | |DW | ||
| - | | | + | |13 ul |
|} | |} | ||
| + | </p> | ||
===Ethanol precipitation=== | ===Ethanol precipitation=== | ||
| + | <p> | ||
For rising concentration of DNA solution which use for Ligation and removing restriction enzyme. | For rising concentration of DNA solution which use for Ligation and removing restriction enzyme. | ||
| - | #Added | + | #Added 3 ul of NaoAc, 1.5 ul of glycogen and 75 ul of 100% ethanol. |
| - | #Centrifuged in | + | #Centrifuged in 14000 rpm, 30 min at 4C. |
#Remove supernatant and added 220ul of 70% ethanol. | #Remove supernatant and added 220ul of 70% ethanol. | ||
| - | #Centrifuged in | + | #Centrifuged in 15000 rpm, 15 min at 4C. |
| - | #Remove supernatant and air drying in room temperature then added | + | #Remove supernatant and air drying in room temperature then added 10 ul of DW. |
| + | </p> | ||
| + | |||
===Ligation=== | ===Ligation=== | ||
| + | <p> | ||
All DNA solutions were digested. | All DNA solutions were digested. | ||
| - | 3A assembly protocol required Ligation reaction should be in total | + | 3A assembly protocol required Ligation reaction should be in total 25 ul solution. |
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
|Ligation Mighty Mix | |Ligation Mighty Mix | ||
| - | |12. | + | |12.5 ul |
|- | |- | ||
|pT7 | |pT7 | ||
| - | | | + | |2 ul |
|- | |- | ||
|RBS | |RBS | ||
| - | | | + | |2 ul |
|- | |- | ||
|pSB1K3 | |pSB1K3 | ||
| - | | | + | |2 ul |
|- | |- | ||
|DW | |DW | ||
| - | |6. | + | |6.5 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |25 ul |
|} | |} | ||
| Line 215: | Line 236: | ||
Withdraw!!!! | Withdraw!!!! | ||
| + | </p> | ||
</div></div> | </div></div> | ||
| Line 226: | Line 248: | ||
<p> | <p> | ||
Transformation for pT7+RBS+pSB1K3 | Transformation for pT7+RBS+pSB1K3 | ||
| - | #Added DNA soltions (Ligation products) | + | #Added DNA soltions (Ligation products) 1 ul to DH5α compitent cell. |
| - | #Stood on ice in | + | #Stood on ice in 30 min. |
| - | #Added | + | #Added 600 ul of LB to transformed DH5α solution. |
| - | #Pre-cultivate in | + | #Pre-cultivate in 2 hrs |
| - | #Splead | + | #Splead 300 ul of LB&DH5α solution to LBK. |
#Cultivated | #Cultivated | ||
</p> | </p> | ||
| Line 238: | Line 260: | ||
mini-prep for Liquid culture product of K346007(Ag43) | mini-prep for Liquid culture product of K346007(Ag43) | ||
#Used FastGene Plasmid Mini Kit(Nippon Genetics) | #Used FastGene Plasmid Mini Kit(Nippon Genetics) | ||
| - | #Elutioned in | + | #Elutioned in 50 ul |
#'''First we eluted in colection tube. then moved in microcentrifuge tube.''' | #'''First we eluted in colection tube. then moved in microcentrifuge tube.''' | ||
</p> | </p> | ||
| Line 245: | Line 267: | ||
[[image:HokkaidoU2012 120708 K346007 miniprep and pt7 rbs.jpg|thumb|mini-prep result (With ligation result of pT7+RBS+pSB1K3)]] | [[image:HokkaidoU2012 120708 K346007 miniprep and pt7 rbs.jpg|thumb|mini-prep result (With ligation result of pT7+RBS+pSB1K3)]] | ||
Erectrophoresis for mini-prep product(Ag43). | Erectrophoresis for mini-prep product(Ag43). | ||
| - | #Prepared 1% Agalose gel and added EtBr then pre-migration in | + | #Prepared 1% Agalose gel and added EtBr then pre-migration in 30 min. |
| - | # | + | #1 ul 1kb ladder, 1.2 ul mini-prep product(1 ul is DNA solution and 0.2 ul is loading dye) added then migtrated. |
</p> | </p> | ||
===Glycerol stock=== | ===Glycerol stock=== | ||
| Line 266: | Line 288: | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | | | + | |48 ul |
|- | |- | ||
|EcoRI | |EcoRI | ||
| - | | | + | |1 ul |
|- | |- | ||
|SpeI | |SpeI | ||
| - | | | + | |1 ul |
|- | |- | ||
|10xH buffer | |10xH buffer | ||
| - | | | + | |6 ul |
|- | |- | ||
DW | DW | ||
| - | | | + | |4 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |60 ul |
|} | |} | ||
*dT(Vector) | *dT(Vector) | ||
| Line 288: | Line 310: | ||
|- | |- | ||
|DNA solution | |DNA solution | ||
| - | | | + | |8 ul |
|- | |- | ||
|EcoRI | |EcoRI | ||
| - | | | + | |1 ul |
|- | |- | ||
|XbaI | |XbaI | ||
| - | | | + | |1 ul |
|- | |- | ||
|10xM buffer | |10xM buffer | ||
| - | | | + | |2 ul |
|- | |- | ||
|DW | |DW | ||
| - | | | + | |8 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |20 ul |
|} | |} | ||
[[image:HokkaidoU2012 120708 dt K346007 digestion umeuchi.jpg|thumb|Digestion result]] | [[image:HokkaidoU2012 120708 dt K346007 digestion umeuchi.jpg|thumb|Digestion result]] | ||
| Line 315: | Line 337: | ||
<p> | <p> | ||
Ethanol precipitation for gel extraction products(K346007(Ag43) and B0015(dT)) | Ethanol precipitation for gel extraction products(K346007(Ag43) and B0015(dT)) | ||
| - | #Added | + | #Added 5 ul of NaoAc , 1.5 ul of glycogen and 125 ul of 100% ethanol to 50 ul DNA solutions. |
| - | #Centrifuged in | + | #Centrifuged in 15000 rpm, 10min at 4C. |
| - | #Remove supernatant and added | + | #Remove supernatant and added 220 ul of 70% ethanol. |
| - | #Centrifuged in | + | #Centrifuged in 15000 rpm, 5 min at 4C. |
| - | #Remove supernatant and air drying in room temperature then added | + | #Remove supernatant and air drying in room temperature then added 10 ul of DW. |
</p> | </p> | ||
===Ligation=== | ===Ligation=== | ||
| Line 328: | Line 350: | ||
|- | |- | ||
|Ligation Mighty Mix | |Ligation Mighty Mix | ||
| - | | | + | |5 ul |
|- | |- | ||
|Insert: Ag43 | |Insert: Ag43 | ||
| - | | | + | |2 ul |
|- | |- | ||
|Vector: dT | |Vector: dT | ||
| - | | | + | |2 ul |
|- | |- | ||
|DW | |DW | ||
| - | | | + | |1 ul |
|- | |- | ||
|Total | |Total | ||
| - | | | + | |10 ul |
|} | |} | ||
Ligation reaction recipe is following. | Ligation reaction recipe is following. | ||
| Line 362: | Line 384: | ||
[[image:HokkaidoU2012 120708 K346007-dT ligation.jpg|thumb|Erectrophoresis result]] | [[image:HokkaidoU2012 120708 K346007-dT ligation.jpg|thumb|Erectrophoresis result]] | ||
Confirmation of succession of ligation. | Confirmation of succession of ligation. | ||
| - | #Prepared 1% Agalose gel and added EtBr then pre-migration in | + | #Prepared 1% Agalose gel and added EtBr then pre-migration in 30 min. |
| - | #Added 1kb ladder, Ligation product( | + | #Added 1kb ladder, Ligation product(1 ul) and digestion products (control:each solutions 1 ul). |
| - | #Migtrated in | + | #Migtrated in 30 min. |
</p> | </p> | ||
===Transformation=== | ===Transformation=== | ||
<p> | <p> | ||
Transformation for K346007(Ag43)+B0015(dT) on pSB1AK3. | Transformation for K346007(Ag43)+B0015(dT) on pSB1AK3. | ||
| - | #Added DNA soltions (Ligation products) | + | #Added DNA soltions (Ligation products) 1 ul to DH5α compitent cell. |
| - | #Stood on ice in | + | #Stood on ice in 30 min. |
| - | #Added | + | #Added 600 ul of LB to transformed DH5α solution. |
| - | #From 700 solution( | + | #From 700 solution(100 ul is DH5α and 600 ul is LB), 100 ul add to 900 ul of LB(x10 solution) |
| - | #Spread | + | #Spread 300 ul from 600(700-100) ul and 1000 ul of LB&DH5α solution to each LBA plates. |
#Cultivated. | #Cultivated. | ||
</p> | </p> | ||
Revision as of 08:03, 6 August 2012
Contents |
July 2nd
On your mark...
July 3rd
Get set...
July 4th
Transformation
Transformation of BBa_B0015(dT), B0034(RBS), I179005(pT7), K346007(Ag43), and K542009(pLacI-RBS-Ag43) in DH5α
- Added each DNA solutions (1 ul) into DH5α comeptent cell and standed on ice in 30 min.
- Cultivated on LBA(dt,RBS,T7) and LBC(Ag43, pLacI-RBS-Ag43). E.coli cultivate in LBC was pre-cultivated in 2 hrs. pT7 was 21 hrs and Others were 20 hrs cultivated.
July 5th
Transformation
K346007(Ag43) was failed to cultivate on LBC plate. Transformation of K346007(Ag43) in DH5α.
- Add DNA solution(1 ul) into DH5α comeptent cell and stand on ice in 30 min.
- Pre-cultivated in 2 hrs.
- Cultivated on LBC in 21hrs.
Single colony isolation
Single colony isolation of BBa_B0015, B0034, I179005 and K542009.
- Picked up one colony.
- Cultivation on LBA(dt,RBS,T7) and LBC(pLacI-RBS-Ag43) in 14 hrs and 30 mins
July 6th
Liquid culture
Liquid culture in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43)
- Picked up two colonies from each plates.
- One colony was resuspended in 1 ml LB (A or C), and other colony was resuspended in 2 ml LB(A or C). 1 ml is for glycerol stocks and 2 ml is for mini-prep.
- 16 hrs Cultivation.
Single colony isolation
- Single colony isolation of K346007(Ag43) on LBC.
July 7th
Liquid culture
Liquid culture in LBC(Ag43).
- Resuspended two colonies from each plates.
- Both of colonies were dipped in 2 ml LBC, and then we cultivate them in 38C.
3A assembly
Assembled pT7, RBS and pSB1C3 by 3A assembly. This 3A assembly is our first try!
mini-prep
- mini-prep of dT,RBS,pT7 and pLacI-RBS-Ag43. We used FastGene Plasmid Mini Kit(Nippon Gene)
- Got 50 ul of DNA solutions.
Glycerol stock
glycerol stocks of dT,RBS,pT7 and pLacI-RBS-Ag43.
- Parts written above were cultivated in LBA(dT,RBS,pT7) and LBC(pLacI-RBS-Ag43) 1 ml in 16 hrs 30 min.
- Add glycerol and Freeze at -80C
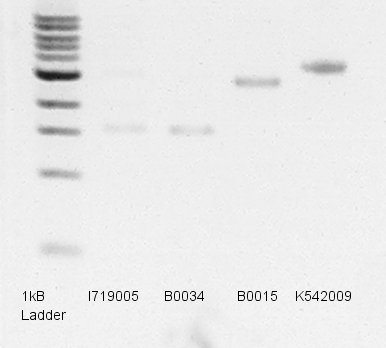
Electrophoresis
Electrophoresis to predict concentration of mini-prep products(dT,RBS,pT7 and pLacI-RBS-Ag43).
- Used 1% agarose gel.
- Added EtBr into TBE buffer and started Pre-migration.
- Migrated 1.2 ul of DNA solutions (1 ul is mini-prep products and 0.2 ul is Loading Dye) in 35 min.
- Took a photograph of 1% agarose gel that finished electrophoresis.
Digestion
Digestion of I719005, B0034 and pSB1K3 Digestion recipe All parts were reacted in 30ul digestion mix.
- I719005(40 ng/ul)
| DNA solution | 12.5 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH Buffer | 3 ul |
| DW | 12.5 ul |
- B0034(40 ng/ul)
| DNA solution | 12.5 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM Buffer | 3 ul |
| DW | 12.5 ul |
- pSB1K3(25 ng/ul)
| DNA solution | 12 ul |
| EcoRI | 1 ul |
| PstI | 1 ul |
| 10xH Buffer | 3 ul |
| DW | 13 ul |
Ethanol precipitation
For rising concentration of DNA solution which use for Ligation and removing restriction enzyme.
- Added 3 ul of NaoAc, 1.5 ul of glycogen and 75 ul of 100% ethanol.
- Centrifuged in 14000 rpm, 30 min at 4C.
- Remove supernatant and added 220ul of 70% ethanol.
- Centrifuged in 15000 rpm, 15 min at 4C.
- Remove supernatant and air drying in room temperature then added 10 ul of DW.
Ligation
All DNA solutions were digested. 3A assembly protocol required Ligation reaction should be in total 25 ul solution.
| Ligation Mighty Mix | 12.5 ul |
| pT7 | 2 ul |
| RBS | 2 ul |
| pSB1K3 | 2 ul |
| DW | 6.5 ul |
| Total | 25 ul |
Ligation reaction recipe was written below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
ligation was finished.
But now pm10:00. 2.5hrs needs for doing transformation. Transformation would finish at am:0:30.
Withdraw!!!!
July 8th
- (pT7 + RBS)
Transformation
Transformation for pT7+RBS+pSB1K3
- Added DNA soltions (Ligation products) 1 ul to DH5α compitent cell.
- Stood on ice in 30 min.
- Added 600 ul of LB to transformed DH5α solution.
- Pre-cultivate in 2 hrs
- Splead 300 ul of LB&DH5α solution to LBK.
- Cultivated
- K346007(Ag43)
mini-prep
mini-prep for Liquid culture product of K346007(Ag43)
- Used FastGene Plasmid Mini Kit(Nippon Genetics)
- Elutioned in 50 ul
- First we eluted in colection tube. then moved in microcentrifuge tube.
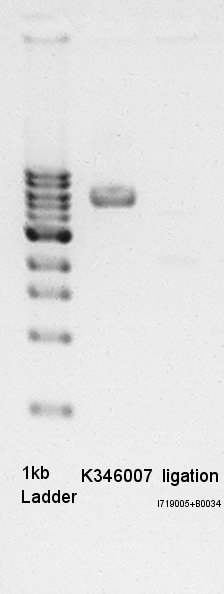
Erectrophoresis
Erectrophoresis for mini-prep product(Ag43).
- Prepared 1% Agalose gel and added EtBr then pre-migration in 30 min.
- 1 ul 1kb ladder, 1.2 ul mini-prep product(1 ul is DNA solution and 0.2 ul is loading dye) added then migtrated.
Glycerol stock
Made glycerol stock of K346007 (Ag43).
- Parts written above were cultivated in LBC.
- Added glycerol and freezed at -80C
- (Ag43 + dT)
Assembling K346007(Ag43) + B0015(dT) with 2-piece assembly(Biobrisk standard assembly)
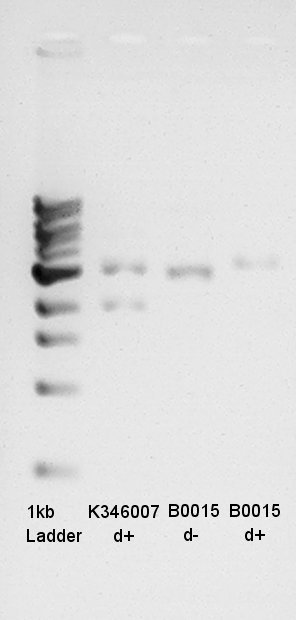
Digestion
Digested Ag43 and dT in solution by recipes Written below. Insert DNA required too much weight and volume(volume was calculated from concentration of DNA mini-prep product)from our calculation. There are no insurance of succession of digestion.
- Ag43(Insert)
| DNA solution | 48 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 6 ul |
| 4 ul | |
| Total | 60 ul |
- dT(Vector)
3318bp(Ag43 + pSB1AK3)
| DNA solution | 8 ul |
| EcoRI | 1 ul |
| XbaI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 8 ul |
| Total | 20 ul |
K346007(Ag43) was 5190bp before digestion (Biobrick prefix + Ag43 + Biobrick suffix + pSB1C3). After digestion, Ag43 and pSB1C3 was separated and became fragments about 3120bp(Ag43) and 2070bp(pSB1C3). Gel image above shows there are two fragments, one fragment is about 2000bp other fragment is 3000bp. Digestion would be succeeded. About Vector(Boo15:dT), the DNA was circular so DNA migrated more far than Linear DNA. d+ (Circular DNA) migrated little more far than d- (Linear DNA). so we think digestion was succeeded.
Ethanol precipitation
Ethanol precipitation for gel extraction products(K346007(Ag43) and B0015(dT))
- Added 5 ul of NaoAc , 1.5 ul of glycogen and 125 ul of 100% ethanol to 50 ul DNA solutions.
- Centrifuged in 15000 rpm, 10min at 4C.
- Remove supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 5 min at 4C.
- Remove supernatant and air drying in room temperature then added 10 ul of DW.
Ligation
All DNA solutions were digested. Used Ligation Mighty Mix(TakaraBio)
| Ligation Mighty Mix | 5 ul |
| Insert: Ag43 | 2 ul |
| Vector: dT | 2 ul |
| DW | 1 ul |
| Total | 10 ul |
Ligation reaction recipe is following.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
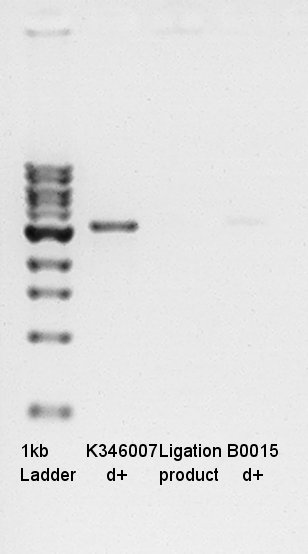
Electrophoresis
Confirmation of succession of ligation.
- Prepared 1% Agalose gel and added EtBr then pre-migration in 30 min.
- Added 1kb ladder, Ligation product(1 ul) and digestion products (control:each solutions 1 ul).
- Migtrated in 30 min.
Transformation
Transformation for K346007(Ag43)+B0015(dT) on pSB1AK3.
- Added DNA soltions (Ligation products) 1 ul to DH5α compitent cell.
- Stood on ice in 30 min.
- Added 600 ul of LB to transformed DH5α solution.
- From 700 solution(100 ul is DH5α and 600 ul is LB), 100 ul add to 900 ul of LB(x10 solution)
- Spread 300 ul from 600(700-100) ul and 1000 ul of LB&DH5α solution to each LBA plates.
- Cultivated.
 "
"