Team:Groningen/Project
From 2012.igem.org








Contents |
Summary
Every year, 1,300,000,000 tons of food are thrown away worldwide. This is one third of the global food production. One of the reasons for this is the fact that best before dates are imprecise. A reliable way of monitoring whether food is spoiled or not could save up to 600 euro per household per year.
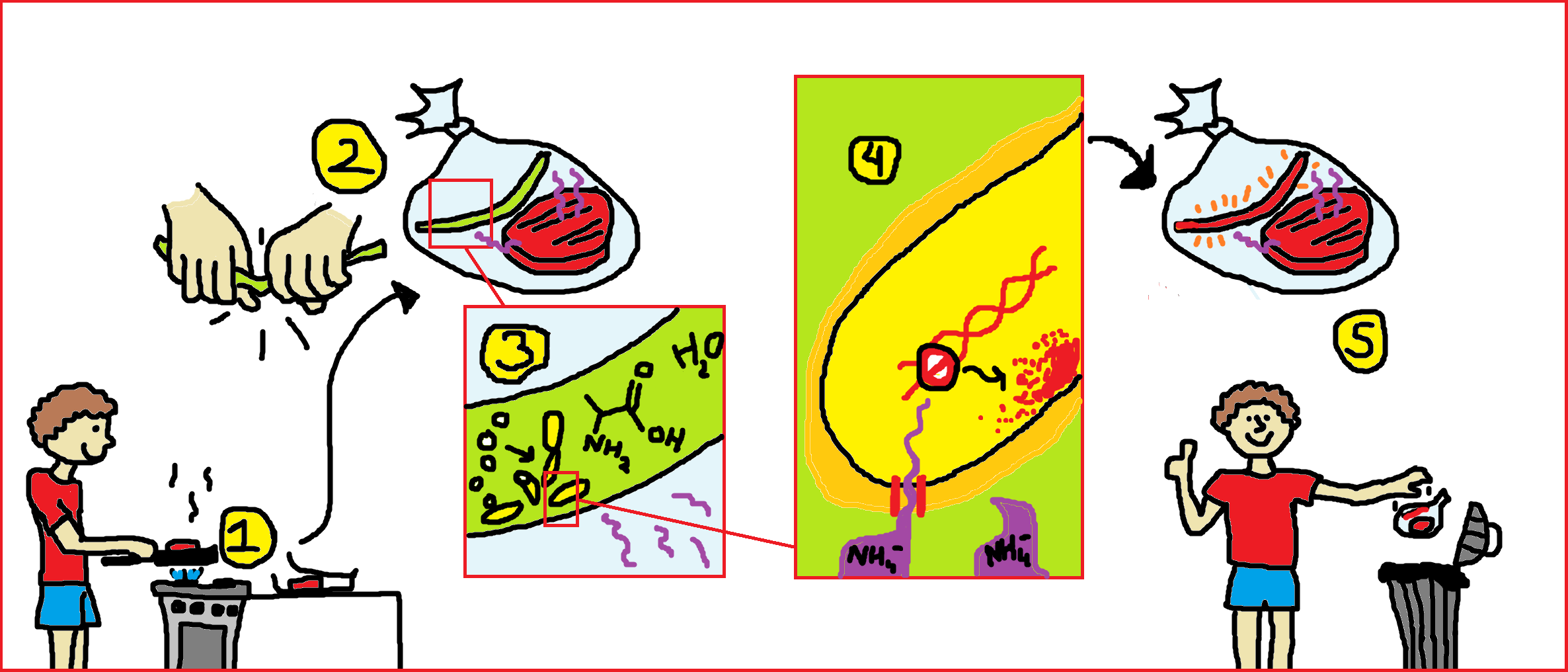
Our goal is to build a system to sense meat spoilage. When a package of meat is saved after opening (1), a closed sticker containing Bacillus subtilis spores is activated by breaking the middle compartment (2,similar to activating a glow-in-the-dark stick), whereby Alanine, and water are mixed with the spores to cause germination (3). When the germinated microbes come in contact with volatiles from rotting meat, a pathway is activated which results to the production of a pigment (4). When this happens, the consumer knows that he has to throw away the food (5) and avoids subsequent food poisoning. The different states of the volatile sensor are explained below.
States
Containment of Bacillus subtilis spores: the sticker
The spores of Bacillus subtilis will be contained in a compartment which is impermeable to bacteria and liquid, but can still let through volatiles. Metabolites, water, and alanine should be separated from the spores by a breakable membrane. Mixing these compounds will cause germination.
The sticker itself is made of a materials that is able to let go gases inside but keeps liquids, spores and bacteria inside and sealed off from the environment, see our safety page. For this purpose TPX polymer would suit the best because it has pores with a size of 0.5um. We want to make a prototype for testing our Bacillus spores.
Germination
Alanine and water can trigger germination of Bacillus subtilis. The modeling team will provide information on the optimal concentrations for germination, and on the germination time.
Activation Pathways
We follow two different strategies to build a pathway in Bacillus subtilis which can react to the presence of rotten meat volatiles. One is using the TnrA pathway, which is normally expressed in absence of ammonium (reference). For the other strategy, we use a transcriptomics approach searching for promoters present in Bacillus reacting to rotten meat volatiles.
First strategy: TnrA pathway
Pre-Activation
- The growth medium contains a low concentration of glutamine (gln), but no ammonia or ammonium (NH4).
- Glutamine synthetase (GS) is activated due to the inadequate level of glutamine, however, it cannot synthesize more gln as it is lacking NH4.
- The low intracellular gln level shifts the ratio of GS to feedback-inhibited GS (FBI-GS) towards a higher level of GS.
- This higher level of GS allows a higher level of TnrA.
- The TnrA represses the alsT promoter.
Activation
- Rotting meat produces NH4.
- NH4 is enters the cell through the NrgAB ammonium transporter.
- GS converts NH4 into gln.
- Gln reaches the concentration required for steady cell growth.
- The ratio of GS to FBI-GS shifts towards a higher level of FBI-GS.
- The newly created FBI-GS binds to TnrA, creating an inactive complex.
- TnrA is unable to repress the alsT promoter.
- No longer repressed, alsT activates along with its positive feedback loop containing the color reporter.
Second strategy: identification of “PBADmeat”
TnrA is a repressor/activator which acts on a broad level. This makes it a tricky candidate for triggering a single reaction. Therefore, an alternative strategy is needed.
To identify our rotten meat volatile reporter (short: PBADmeat), we performed an transcriptome analysis of Bacillus subtilis subjected to rotten meat (target) and to fresh meat (control) volatiles. Analysis of the differential expression between these two situations can lead to candidate promoters for our construct.
For more information on the technique and some preliminary results, see the Wetwork page.
Color reporter
As a reporter, we chose to use a pigment. The big advantage of using a pigment over another type of reporter (for instance, GFP or an electrical signal) is that it can be detected without any equipment. A big drawback however, is the production speed of pigment.
To make Bacillus subtilis produce more pigment, we plan to build in a positive feedback loop. Different feedback loops are available in the Registry of Standard Biological Parts (for instance PLux/LuxR and pRE/CII). We plan to characterize and - if necessary - improve one for our construct.
We will start with using the E.chromi pigments from the iGEM team of Cambrige, 2008. These pigments (violacein and lycopene) are produced strongly in E. coli and are visible by eye at even low optical densities. To test if our construct principally works, however, we will use GFP.
As an alternative for the pigments, we we're provided with chromoproteins from Uppsala 2011. These biobricks were send to us in agar stabs and produce yellow or blue color. We might be able to test if they work in Bacillus.
Death
To make sure no live GMO will get into the environment, the death of Bacillus should be reached:
- The extracellular nutrients are depleted.
- Color reporter builds to toxic levels.
Construct
The standard construct is a combination of biobricks.
- Backbone:Available in registry: BBa_K090403 (Backbone) - Single copy shuttle vector. Other option: build our own well-characterized backbone for our chassis.
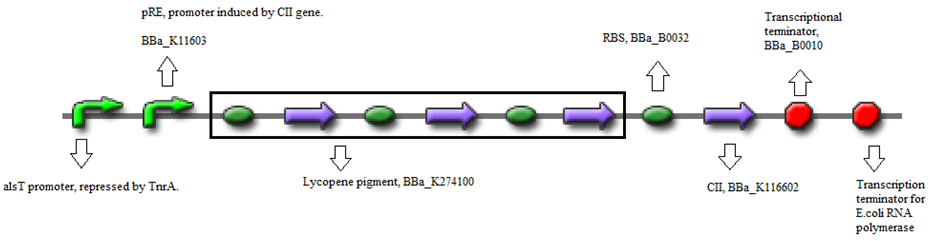
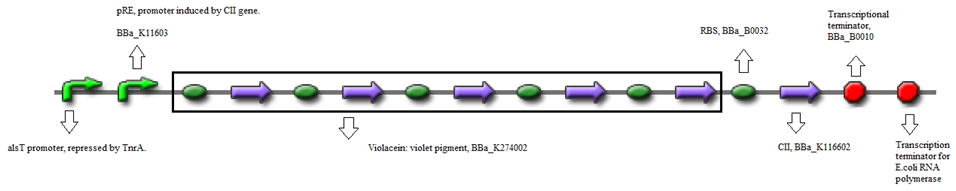
- BBa_K116632+BBa_K116602 (Positive feedback loop) - Uses the pRe promoter regulated by C11 and terminated by B0010. Other options: BBa_J37019;BBa_K116639; BBaK116609.
- BBa_K274100 (Reporter) - Modified carotenoid pathway to produce red pigment.
- Or, using BBa_K274002 (Reporter) - Violet pigment producer</FONT>
Modeling-Labwork Cooperation
Unlike previous years, there is a direct link between theory and practice.
The modeling team of one will provide to the lab:
- Creation, and critical analysis, of the reaction pathways from literature.
- Identification
- of the metabolites necessary to support growth. (Done: D-Glucose, Water, Glutamine, Potassium)
- of the metabolites required for the reporter.
- of the metabolites required for germination.
- of the conditions for sporulation.
- Quantification
- Rates of nutrient consumption versus temperature and biomass.
- Amount of O2 required, and the corresponding volume of air.
- Plot of growth rate vs. temperature.
- Plot of biomass amount vs. time
- Superimpose curves with different starting medium concentration.
- Indicate the points at which sporulation occurs.
The lab will provide to the modeling team:
- TnrA concentration as a function of extracellular glutamine concentration.
- Growth rate as a function of extracellular glutamine and potassium concentrations.
- Rates of volatile production as a function of meat type and volume.
Limitations due to time constraints
- We chose to make our construct at room temperature (37 degrees). If this project leads to a proof of concept, a psychrotrophic bacterium like Bacillus cereus could be used as a chassis instead of Bacillus subtilis. This bacterium is however a much harder to engineer and, unlike Bacillus subtilis, harmful.
Standard Operating Protocols
In an effort to apply business concepts to the iGEM project we have agreed to conduct the project according to the following protocols. The specifics of these sections (such as the actual protocols or equipment listing) are contained within the SOP binder in the laboratory. This binder will be digitized for next year’s iGEM teams.
General
- How to set up an experiment.
- Fully describe the experiment in a document before it is scheduled to be performed (document is described in point 2).
- After the experiment is documented, it should be reviewed by at least one other iGEM member.
- Upon completion of the experiment there should be a short discussion/interpretation of the results and a short outlook for subsequent experiments.
- Experiment Template
- Insert outline of the template here
- Acronyms.
- Each iGEM member is assigned an acronym: e.g. Marius Uebel will be MU.
- Data management.
- Each filename should contain the original creator, date, and type of file. E.g. MU_20120412_igem12_sop_proposal.doc
- Ordering of material.
- A single person should be responsible for ordering, this prevents multiple orders of an item and a more controlled inventory.
Lab Equipment and Materials
- Equipment.
- Every piece of lab equipment should be accompanied by the manufacturer’s manual and a short how-to manual.
- Overview and location of all our equipment should be written down in this part
- Protocols
- All lab protocols should be consolidated into a central location in the lab in print form.
Methodology
- Culture
- Contains general information and cultivation requirements of the chassis
- Assay
- Short overview of any assay kits changes undertaken to suit the actual experiment.
- All information on original assays
 "
"