Team:Frankfurt/New Yeast RFC
From 2012.igem.org
(→Introduction) |
|||
| Line 24: | Line 24: | ||
==Design of DNA fragments for gap repair cloning== | ==Design of DNA fragments for gap repair cloning== | ||
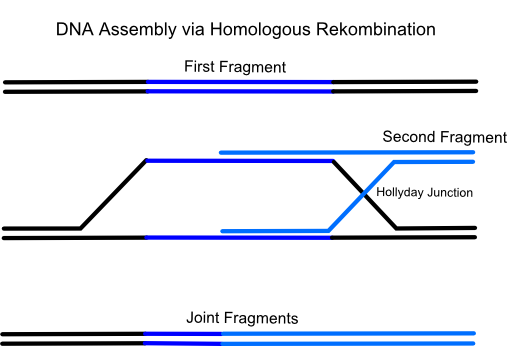
| - | [[Image:GapRepairPictureFrankfurt.png|400px|thumb]]The idea of the method is to transform a series of linear, successive DNA fragments into one yeast cell. The linear fragments have open blunt ends like they occur after a double strand break. If a homologous sequence is available it will be treated like a genomic double strand break and homologous recombination takes place. When the successive DNA fragments are designed in a specific way which includes large sequence overlaps (overall app. 40 bp) to the respectively following fragment yeast will recombinate them together. <br> | + | [[Image:GapRepairPictureFrankfurt.png|400px|thumb]]The idea of the method is to transform a series of linear, successive DNA fragments into one yeast cell. The linear fragments have open blunt ends like they occur after a double strand break. If a homologous sequence is available it will be treated like a genomic double strand break and homologous recombination takes place. When the successive DNA fragments are designed in a specific way which includes large sequence overlaps (overall app. 40 bp) to the respectively following fragment yeast will recombinate them together. <br><br> |
| - | For the formation of a cloning vector the first fragment is a yeast-''E.coli'' shuttle plasmid which is linearized by an appropriate restriction digest. A shuttle plasmid is a plasmid which is stable both in yeast and in ''Escherichia coli''. The first fragment of the insert has to possess an homologous overlap to both the wished insertion site on the plasmid and to the beginning of the second fragment. The end of the second fragment has to possess an overlap to the beginning of the third one and so on. At least the end of the last fragment of the insert again has to possess an overlap homologous to the second insertion site on the plasmid. <br> | + | For the formation of a cloning vector the first fragment is a yeast-''E.coli'' shuttle plasmid which is linearized by an appropriate restriction digest. A shuttle plasmid is a plasmid which is stable both in yeast and in ''Escherichia coli''. The first fragment of the insert has to possess an homologous overlap to both the wished insertion site on the plasmid and to the beginning of the second fragment. The end of the second fragment has to possess an overlap to the beginning of the third one and so on. At least the end of the last fragment of the insert again has to possess an overlap homologous to the second insertion site on the plasmid. <br><br> |
At the lab of our instructor up to eighteen single fragments were assembled in a single transformation. Another advantage of the method is that no scars are left between the inserted fragments. Assembly of fragments to joint genes is possible. Restriction enzymes only have to be used once for linearization of the shuttle plasmid. | At the lab of our instructor up to eighteen single fragments were assembled in a single transformation. Another advantage of the method is that no scars are left between the inserted fragments. Assembly of fragments to joint genes is possible. Restriction enzymes only have to be used once for linearization of the shuttle plasmid. | ||
| Line 31: | Line 31: | ||
{|width="100%" align="center" | {|width="100%" align="center" | ||
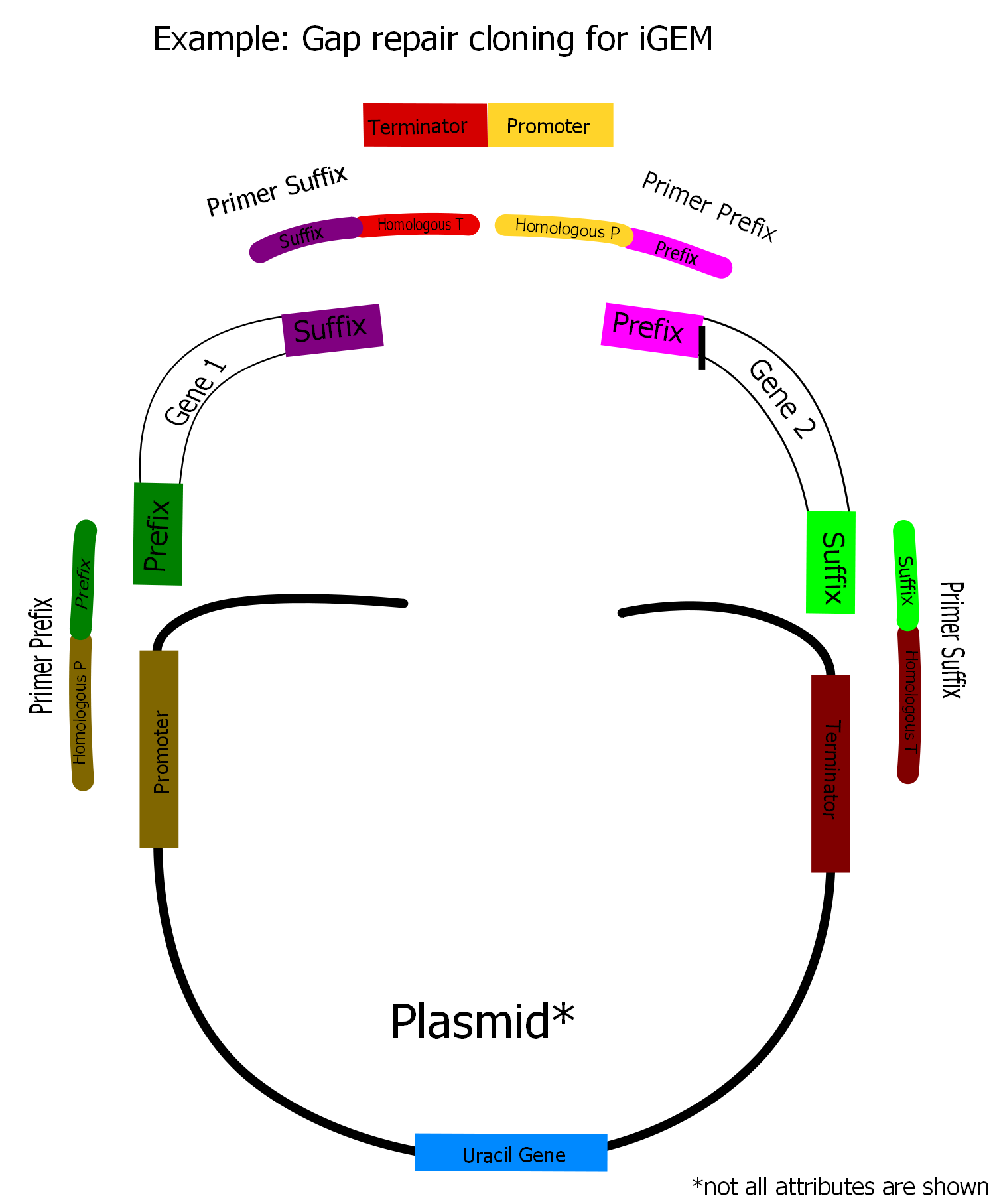
[[Image:Unser_Vektor.png|400px|thumb|The overlap sequences at the beginning and the end of each fragment are 20 bp long and homologous to the respectively previous and subsequent fragment. The overlaps to promoter and terminator on the plasmid are longer (40 bp) because the plasmid does not contain any overlaps to the first and last insert fragment. Thus the homologous region between every fragmentpair is always 40 pb long.]] | [[Image:Unser_Vektor.png|400px|thumb|The overlap sequences at the beginning and the end of each fragment are 20 bp long and homologous to the respectively previous and subsequent fragment. The overlaps to promoter and terminator on the plasmid are longer (40 bp) because the plasmid does not contain any overlaps to the first and last insert fragment. Thus the homologous region between every fragmentpair is always 40 pb long.]] | ||
| - | In our project we have constructed a plasmid for overexpression of three genes of the [[Team:Frankfurt/Project|Mevalonate]] pathway. Therefore we used synthesized fragments of the mentioned three genes, two promoters, two terminators (both from yeast) and a yeast expression plasmid which already contained one promoter, one terminator and a gene for uracil synthesis. Moreover we used a mutant strain where this uracil gene is deleted. So we needed the uracil gene on the plasmid as a marker for selection of the transformants. Via PCR we assembled appropriate homologous overlaps to our fragments using primers containing these overlaps. By that we were able to arrange our fragments in the order shown in the picture on the right. <br> | + | In our project we have constructed a plasmid for overexpression of three genes of the [[Team:Frankfurt/Project|Mevalonate]] pathway. Therefore we used synthesized fragments of the mentioned three genes, two promoters, two terminators (both from yeast) and a yeast expression plasmid which already contained one promoter, one terminator and a gene for uracil synthesis. Moreover we used a mutant strain where this uracil gene is deleted. So we needed the uracil gene on the plasmid as a marker for selection of the transformants. Via PCR we assembled appropriate homologous overlaps to our fragments using primers containing these overlaps. By that we were able to arrange our fragments in the order shown in the picture on the right. <br><br> |
| - | The DNA of all fragments is transformed together into competent yeast cells. The transformants were selected on synthetic medium lacking from uracil (only transformants should be able to grow). After preparing the plasmid from the transfomed yeast clones it can be transformed into ''Escherichia coli'' and gained from there in large amounts for further use. | + | The DNA of all fragments is transformed together into competent yeast cells. The transformants were selected on synthetic medium lacking from uracil (only transformants should be able to grow). After preparing the plasmid from the transfomed yeast clones it can be transformed into ''Escherichia coli'' and gained from there in large amounts for further use.<br><br> |
| + | The DNA sequence of the complete expression vectore is shown in the following file: | ||
| + | [[DNA_Sequence_of_Plasmid_for_Overexpression_of_Mevalonat_Pathway.txt|file]] | ||
|} | |} | ||
| - | + | ||
==Gap repair cloning for iGEM== | ==Gap repair cloning for iGEM== | ||
Revision as of 20:22, 26 September 2012
| Home | Team | Project | Organisms | New Yeast RFC | Notebook | Registered Parts | Modeling | Safety | Attributions | Official Team Profile |
|---|
Contents |
The benefit of vector assembly in yeast
Introduction
iGEM Team Frankfurt 2012 successfully used a relatively new methode for vector assembly in yeast called Gap Repair Cloning. It is a more and more established methode for efficient, fast and error-free construction of plasmids based on the homologous recombination system of Saccharomyces cerevisiae (common yeast). Naturally yeast uses this process to repair DNA double strand breaks which are one of the most dangerous and life-threatening damages of the DNA for a cell. Therefor this eucaryotic microorganism has developed a few enzymes which have the ability to repair a broken DNA double strand by pairing it with a very similiar DNA region (typically on the homologous chromosome).
Using gap repair cloning a series of linear, successive DNA fragments with homologous overlaps to the respectively following fragment can be transformed in only one step into a yeast cell. After that the micoorganism recombines all fragments in the predetermined, specific order to the final targeting vector. The advantage is that up to eighteen and more successive DNA fragments can be assembled in a single transformation. Also only one restriction enzyme for linearization of the plasmid is needed.
For these reasons iGEM Team Frankfurt thought that gap repair cloning is usefull tool for next iGEM generations. Therefore we developed a standardized methode that describes a new way of assembling BioBrick devices in a desired order to a targeting plasmid using homologue recombination system of yeast. It is called Yeast BioBrick Assembly (YBA). YBA standard only needs one restriction enzyme and a standardized selection of primers and promotors/termintors. It is a continuation of the BioBrick standard and compartible with all BBF RFC 10 parts. Additionally it can be adapted by specific primer design to all other BioBrick standards. In the following we focuse on assembly of yeast expression vectors by using YBA methode. However it also can be used for E.coli vector design or assembly.
Homologue Recombination System of Yeast
There are many endogenous and exogenous factors (for example reactive oxygen-species, ionizing radiation, chemicals and failing of DNA binding enzymes (e.g. collapsed replication forks)) which causes DNA double strand breaks. For the cell this is the most dangerous DNA damage because even if it occurs in rather unimportant regions the cell will not survive the next cell cycle. That's the reason why yeast possesses highly active enzymes which have the ability to repair a broken double strand by pairing it with a very similiar DNA region (typically on the homologous chromosome). This process is called homologous recombination. Using the gap repair method this natural process can be exploited for the construction of large cloning vectors in yeast.
Design of DNA fragments for gap repair cloning
The idea of the method is to transform a series of linear, successive DNA fragments into one yeast cell. The linear fragments have open blunt ends like they occur after a double strand break. If a homologous sequence is available it will be treated like a genomic double strand break and homologous recombination takes place. When the successive DNA fragments are designed in a specific way which includes large sequence overlaps (overall app. 40 bp) to the respectively following fragment yeast will recombinate them together.For the formation of a cloning vector the first fragment is a yeast-E.coli shuttle plasmid which is linearized by an appropriate restriction digest. A shuttle plasmid is a plasmid which is stable both in yeast and in Escherichia coli. The first fragment of the insert has to possess an homologous overlap to both the wished insertion site on the plasmid and to the beginning of the second fragment. The end of the second fragment has to possess an overlap to the beginning of the third one and so on. At least the end of the last fragment of the insert again has to possess an overlap homologous to the second insertion site on the plasmid.
At the lab of our instructor up to eighteen single fragments were assembled in a single transformation. Another advantage of the method is that no scars are left between the inserted fragments. Assembly of fragments to joint genes is possible. Restriction enzymes only have to be used once for linearization of the shuttle plasmid.
Example from our project
 "
"