Team:Evry/Protocols
From 2012.igem.org
Chr.karine (Talk | contribs) |
Chr.karine (Talk | contribs) |
||
| Line 2: | Line 2: | ||
---- | ---- | ||
| + | <h1>Midi-prep</h1> | ||
| + | <i>Protocol from QIAGEN plasmid purification handbook</i><br> | ||
| + | Pre-chill Buffer P3 at 4°C <br> | ||
| + | <b>1.</b>Pick a single colony from a freshly streaked selective plate and inoculate a starter | ||
| + | culture of 2–5 ml LB medium containing the appropriate selective antibiotic. Incubate | ||
| + | for approx. 8 h at 37°C with vigorous shaking (approx. 300 rpm). | ||
| + | Use a tube or flask with a volume of at least 4 times the volume of the culture. | ||
| + | <b>2.</b>Dilute the starter culture 1/500 to 1/1000 into selective LB medium. For high-copy | ||
| + | plasmids, inoculate ▲ 25 ml or ● 100 ml medium with ▲ 25–50 μl or | ||
| + | ● 100–200 μl of starter culture. For low-copy plasmids, inoculate ▲ 100 ml or | ||
| + | ● 500 ml medium with ▲ 100–200 μl or ● 250–500 μl of starter culture. Grow at | ||
| + | 37°C for 12–16 h with vigorous shaking (approx. 300 rpm). | ||
| + | Use a flask or vessel with a volume of at least 4 times the volume of the culture. The | ||
| + | culture should reach a cell density of approximately 3–4 x 109 cells per milliliter, | ||
| + | which typically corresponds to a pellet wet weight of approximately 3 g/liter | ||
| + | medium. | ||
| + | 3. Harvest the bacterial cells by centrifugation at 6000 x g for 15 min at 4°C. | ||
| + | ƒ If you wish to stop the protocol and continue later, freeze the cell pellets at –20°C. | ||
| + | 4. Resuspend the bacterial pellet in ▲ 4 ml or ● 10 ml Buffer P1. | ||
| + | For efficient lysis it is important to use a vessel that is large enough to allow complete | ||
| + | mixing of the lysis buffers. Ensure that RNase A has been added to Buffer P1. | ||
| + | If LyseBlue reagent has been added to Buffer P1, vigorously shake the buffer bottle | ||
| + | before use to ensure LyseBlue particles are completely resuspended. The bacteria | ||
| + | should be resuspended completely by vortexing or pipetting up and down until no | ||
| + | cell clumps remain. | ||
| + | |||
| + | |||
<h1>PCR with Phusion High-Fidelity DNA Polymerase</h1> | <h1>PCR with Phusion High-Fidelity DNA Polymerase</h1> | ||
Revision as of 15:17, 8 August 2012
Contents |
Midi-prep
Protocol from QIAGEN plasmid purification handbook
Pre-chill Buffer P3 at 4°C
1.Pick a single colony from a freshly streaked selective plate and inoculate a starter
culture of 2–5 ml LB medium containing the appropriate selective antibiotic. Incubate
for approx. 8 h at 37°C with vigorous shaking (approx. 300 rpm).
Use a tube or flask with a volume of at least 4 times the volume of the culture.
2.Dilute the starter culture 1/500 to 1/1000 into selective LB medium. For high-copy
plasmids, inoculate ▲ 25 ml or ● 100 ml medium with ▲ 25–50 μl or
● 100–200 μl of starter culture. For low-copy plasmids, inoculate ▲ 100 ml or
● 500 ml medium with ▲ 100–200 μl or ● 250–500 μl of starter culture. Grow at
37°C for 12–16 h with vigorous shaking (approx. 300 rpm).
Use a flask or vessel with a volume of at least 4 times the volume of the culture. The
culture should reach a cell density of approximately 3–4 x 109 cells per milliliter,
which typically corresponds to a pellet wet weight of approximately 3 g/liter
medium.
3. Harvest the bacterial cells by centrifugation at 6000 x g for 15 min at 4°C.
ƒ If you wish to stop the protocol and continue later, freeze the cell pellets at –20°C.
4. Resuspend the bacterial pellet in ▲ 4 ml or ● 10 ml Buffer P1.
For efficient lysis it is important to use a vessel that is large enough to allow complete
mixing of the lysis buffers. Ensure that RNase A has been added to Buffer P1.
If LyseBlue reagent has been added to Buffer P1, vigorously shake the buffer bottle
before use to ensure LyseBlue particles are completely resuspended. The bacteria
should be resuspended completely by vortexing or pipetting up and down until no
cell clumps remain.
PCR with Phusion High-Fidelity DNA Polymerase
Tube preparation
Put items in this order:
| Component | 50µl reaction | Comments |
| H2O | 32 | |
| 5x Phusion HF Buffer | 10 | |
| 10mM dNTPs | 1 | |
| Primer FW | 2 | Primers have to be at 10µM |
| Primer RV | 2 | Primers have to be at 10µM |
| Template DNA | 1 | |
| DMSO (optional) | 1,5 | recommended for GC-rich amplicons < 20kb |
| Phusion DNA polymerase | 0,5 |
Cycling instructions
| Cycle step | Temperature | Time | Cycles |
| Initial denaturation | 98°C | 4min | 1 |
| Denaturation | 98°C | 20s | 30 |
| Annealing | Lower Tm of primers | 30s | |
| Extension | 72°C | 30S/kb | |
| Final extension | 72°C | 10min | 1 |
| 4°C | hold |
Préparation of LB medium and LB Agar:
=> LB Agar :
-18,5g LB Agar
-300ml H2O
=> LB medium : -6g LB broth -300ml de H2O
Autoclaved at 250°C
Gel extraction
1. Excise the DNA fragment from the agarose gel with a clean, sharp scalpel under UV light.
2. Weight the gel slice in a colorless tube. Add 3 volumes of Buffer QG to 1 volume of gel.
3. Incubate at 50°C for 10 min, until the gel slice has completely dissolved.
4. The color of the mixture have to be yellow, otherwise add 10 µl of 3M sodium acetate.
5. Ass 1 gel volume of isopropanol to the sample and mix.
6. Place a spin column in a provided 2 ml collection tube.
7. To bind DNA, apply the sample to the column and centrifugat for 1 min. Discard the flow-through and place the column back into the same tube.
8. To wash, add 0,75 ml of buffer PE to the column and centrifugate for 1 min.Discard the flow-through and place the column back into the same tube.
9. Centrifugate the column in a 2 ml Collection tube for 1 min 17,900xg (13,000 rpm).
10. Place the column into a clean 1,5 ml microcentrifuge tube.
11. to elute DNA, add 15 µl of water to the column and centrifugate for 1 min.
In vitro fecondation of Xenopus tropicalis
2 or 3 days before the IVF
Pre-injection of frogs (males et females)
100µl of hCG (0,1UI/µl, do a dilution by 10 of the commercial solution)
D day
Test the injector before beginning
Inject the 100µl hCG (1UI/µl) in the morning
Isolate the animals : an animal by box
Prepare :
Timer
Methylcellulose (put at room temperature)
Boxes special for injections
Materiel of injection
Capillaires ad hoc with oil
Solutions to inject (add 1µl of blue or red of Nile)
Solution L15/FCS :
- boxes (1/femelle) of Pétri (d=10 cm) lined of L15/FCS (500µl)
- 2 tubes 1,5mL with 250µl of L15/FCS (one for each testicle)
Pestles in plastic cleaned to grind the testicles
MMR 0,1X
MMR 0,05X
MS-222 0,2%
Euthanasia of the male
Put the male in a vat with MS-222 0,2% and look forward to his non response of pinches. Put it out and process of the dissection to extract the two testicles.
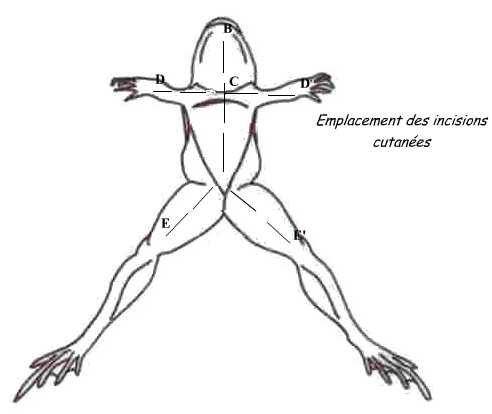
For that, dispose the animal: the dorsal face against the dissection board. Pinch the skin in the middle of the belly and do a small incision with scissors; from this point, perform skin incisions shown in the diagram.
Fold the flaps and skin incision in the muscular wall avoiding severing the abdominal vein to avoid hemorrhage. Both gonads are located in the ventral face of kidneys.
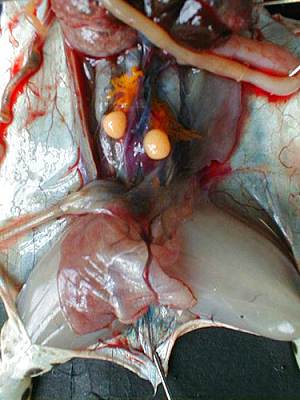
Both gonads are located in the ventral surface of the kidneys, they cover more or less and in corpora lutea (fat mass).By extracting them from the abdominal cavity, we can extract the testicles attached to them. The male's testicles are two oval organs, smooth, yellow-pale as can be seen in the photograph below.
Roll the testicles on kitchen paper to remove blood and blood vessels. Dip each testicle in a tube of 1,5mL with 250µl of L15/FCS. Keep tubes on ice. Crush the testicles with a pestle until obtaining a homogeneous solution. Add 250µl of L15/FCS, mix with a P1000 always on ice.
Lay eggs of females
Spawn females (who have already started laying alone) in Petri dishes previously coated with L15/FCS (gently massage into their flanks).
In vitro fecondation
With a pasteur pipette in plastic, remove as much liquid in the Petri dish prior the IVF. Fil 50µl/500eggs of sperm solution on the aggs with P1000 and mix with a cone to evenly distribute the sperm on the eggs. Let the sperm fix on the eggs during 3-5 min.
Activate the spermatozoid by recovering the eggs of MMR 0,05X to allow the fecundation.
Degangage
15 min after fecondation activation, processing of the degangage.
For that, prepare cysteine 2%, pH 7,5-8 :
- Weight 2g of cysteine powder, transfert if in 80 mL of MMR 0,1% and put it in a becher under magnetic agitation. Mesure pH et equilibrate by adding NaOH 1M.
- Complete with MMR 0,1% to 100mL.
Remove MMR 0,05% from the petri box containing the eggs. Add the cysteine and agitate the box by rotation for 2 min.
Transfere eggs in a 100ml pot (red plug) and continue to agitate by rotation for 5-8 min. check that the embryos seem to touch each other to validate the degangage.
Rinse 3 times with MMR 0,1X.
Attention : during the diferent phases of washing, ensure that the eggs are always in liquid.
Add some methylcellulose to eggs to retard the developpement and facilitate the injection.
Place eggs (50-100) in petri boxes special for injection (lined with a small mesh) and inject.
After injections, the embryos are transfered in MMR 0,1% in small petri boxes lined in the background with agarose. Boxes are incubated at 21°C.
 "
"