Team:Evry/Data
From 2012.igem.org
| (69 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Evry/template_v1}} | {{:Team:Evry/template_v1}} | ||
| + | <html> | ||
| - | + | <h1>Data page: Summary of our summer work</h1> | |
| - | + | ||
| - | + | <br /> | |
| + | <center><img src="https://static.igem.org/mediawiki/2012/c/cd/Frog_tube.png"></center> | ||
| + | <br /> | ||
| - | + | <h2>French Froggies : New Xenopus plasmids for creating multicellular systems</h2> | |
| - | + | ||
| - | + | <center><img src="https://static.igem.org/mediawiki/2012/c/cc/Plasmid_backbone.png" alt="Image unavailable" width="550px" /></center> | |
| + | <p>We developed and submitted to the registry 2 new plasmid backbones for creating multicellular synthetic systems and the corresponding biobricked promoters<p> | ||
| + | <ul> | ||
| + | <li>The pSC2+ with the CMV promoter: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812000">K812000</a></li> | ||
| + | <li>The pSC2+ with pElastase promoter (pancreas specific): <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812200">BBa_K812200</a></li> | ||
| + | <li>And the Biobricked pElastase alones: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812233">BBa_K812233</a></li> | ||
| + | <li>And with a fluorescent citrine to characterize the promoter pElastase in the frog: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812331"></a></li> | ||
| + | <li>We also submitted a heat shock Xenopus promoter: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812331">BBa_K812331</a></li> | ||
| + | </ul> | ||
| - | + | <h2>A serie of new eukaryotic reporters ready for expression</h2> | |
| + | </html> | ||
| + | [[File:Sélection_168.png|thumb|right|Expression of the sfGFP in pCS2+ microinjected in a tadpole]] | ||
| + | <html> | ||
| + | <p>We also prepared a series of fluorescent reporters with a kozak sequence for high expression in eukaryotes.</p> | ||
| - | + | <ul> | |
| + | <li>A kozak-citrine in pSB1C3: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812030">K812030</a></li> | ||
| + | <li>A kozak-sfGFP in pSB1C3: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812031">K812031</a></li> | ||
| + | <li>A kozak-mCFP in pSB1C3: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812032">K812032</a></li> | ||
| + | </ul> | ||
| + | <p>And provided them directely cloned in pSC2+ for direct expression in Xenopus or chicken:</p> | ||
| + | <ul> | ||
| + | <li>The Citrine in pSC2+: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812130">K812130</a></li> | ||
| + | <li>The mCFP in pSC2+: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812132">K812132</a></li> | ||
| + | <li>The sfGFP in pSC2+: <a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812133">K812133</a></li> | ||
| + | </ul> | ||
| + | <p>All these plasmids can be directly injected in to the <i>Xenopus</i> embryo after a miniprep, and are designed for rapid testing of multicellular devices. They contain all sequences necessary for a tissue specific expression, as well as debugging tools. Several plasmids can be co-injected, allowing ratio adjustment and linking the system do modeling.Once the system is tested and debugged, the final system can be implemented by chromosomal insertion. </p> | ||
| - | |||
| + | <h3>Characterization</h3> | ||
| + | </br> | ||
| + | The information on characterized part can be found here: <a href="https://2012.igem.org/Team:Evry/FrenchFrog">FrenchFrog</a> | ||
| - | + | <h2>The auxin system</h2> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <h3>Auxin production device</h3> | |
| - | + | <p> | |
| - | + | We adapted the auxin production module created by the Imperial College 2011 team for it to be used in eukaryotes. We made a system designed for coinjection, and a system which works with a single “operon” using pep2A. This is a short self-cleaving peptide, which will cut a nascent peptide chain to make 2 proteins, insuring stochiometry of the products in the cell. These auxin production devices are designed to communicate through the organism with the auxin reception device.</p> | |
| - | < | + | |
| - | <script type="text/javascript">writeFooter()</script> | + | |
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:502px;"><a href="/DEGRON_PART_1_.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/4/4b/DEGRON_PART_1_.jpg" width="500" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/DEGRON_PART_1_.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 3: Schematic representation of the Auxin two-step production cassette.</div></div></div></div> | ||
| + | |||
| + | <p> <ul> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812020">K812020: </a>IAAH enzyme that catalyzes the second and last step towards auxin production | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812021">K812021: </a>IAAM enzyme that catalyses the transformation of Tryptophan to Inodle-3-acetamide | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812014">K812014: </a>Auxin production cassette containg both enzymes above which is monocystronic | ||
| + | </ul> | ||
| + | </p> | ||
| + | |||
| + | <h3>Auxin reception device</h3> | ||
| + | <p> | ||
| + | The auxin reception device is composed of two components : Tir1, an auxin dependant ubiquitin ligase, and AID – a peptide that is recognized and ubiquitinated specifically by Tir1, when auxin is present. By fusing AID to any protein, this protein will be degraded along with the AID tag when auxin is added to the medium.</p> | ||
| + | |||
| + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:502px;"><a href="/File:Degron_part2.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2012/c/c4/Degron_part2.jpg" width="500" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Degron_part2.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 2: schemetic representation of the Auxin-receiver system</div></div></div></div> | ||
| + | |||
| + | <p> | ||
| + | <ul> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812010">K812010:</a> GFP fusion with the ubuquitinase E3 OsTirI recoginition domain which a main part of the degron system | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812012">K812012:</a> OsTirI Ubiquitinase E3 for AID tagged protein degradation in the presence of auxin | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812013">K812013:</a> The Auxin reciever system golden gate-assembled containing GFP-AID OsTirI polysistronic system for auxin detection in tadpole | ||
| + | </ul> | ||
| + | </p> | ||
| + | |||
| + | |||
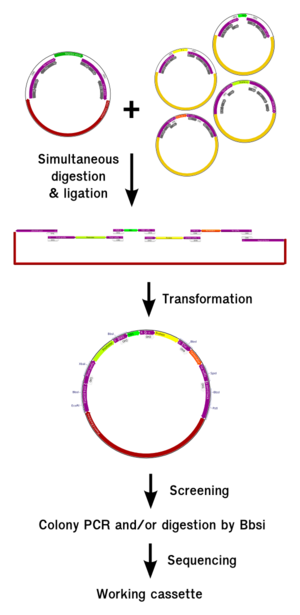
| + | <h2>Goldenbricks</h2> | ||
| + | |||
| + | <h3>Summary</h3> | ||
| + | |||
| + | <div class="thumb tright"><div class="thumbinner" style="width:302px;"><a href="/File:GOldenbrick_global_thumb.png" class="image"><img alt="" src="/wiki/images/thumb/f/f3/GOldenbrick_global_thumb.png/300px-GOldenbrick_global_thumb.png" width="300" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:GOldenbrick_global_thumb.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Fig 4: Summary of the GoldenBrick procedure</div></div></div> | ||
| + | |||
| + | |||
| + | <p>The GoldenBrick is a new assembly method for the partsregistry. If you want to know more, <a href="https://2012.igem.org/Team:Evry/GB">see this page.</a></p> | ||
| + | |||
| + | <h3>Our favourites parts</h3> | ||
| + | |||
| + | <ul> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812050">K812050:</a> A GoldenBricked version of pSB1C3 with J04450 as negative cloning control</li> | ||
| + | <li><a href=="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812051">K812051:</a> A GoldenBricked version of pSB1K3 with J04450 as negative cloning control</li> | ||
| + | </ul> | ||
| + | |||
| + | <h3>The other parts we have created</h3> | ||
| + | |||
| + | <ul> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812050">K812050:</a> A GoldenBricked version of pSB1C3 with J04450 as negative cloning control</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812051">K812051:</a> A GoldenBricked version of pSB1K3 with J04450 as negative cloning control</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812053">K812053:</a> A GoldenBricked version of the strong RBS B0034</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812054">K812054:</a> A GoldenBricked version of the RFP E1010</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812055">K812055:</a> A GoldenBricked version of the terminator B0015</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812056">K812056:</a> A GoldenBricked version of the pLac R0010 promoter</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812057">K812057:</a> A GoldenBricked of an sfGFP protein</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812058">K812058:</a> A GoldenBricked of medium strenght RBS J61107</li> | ||
| + | <li><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K812058">K812059:</a> A GoldenBricked of week RBS J61117</li> | ||
| + | </ul> | ||
| + | |||
| + | <!-- <p>say we are testing them</p> --> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <script type="text/javascript">writeFooter()</script> | ||
</html> | </html> | ||
Latest revision as of 03:40, 27 September 2012
Data page: Summary of our summer work

French Froggies : New Xenopus plasmids for creating multicellular systems

We developed and submitted to the registry 2 new plasmid backbones for creating multicellular synthetic systems and the corresponding biobricked promoters
- The pSC2+ with the CMV promoter: K812000
- The pSC2+ with pElastase promoter (pancreas specific): BBa_K812200
- And the Biobricked pElastase alones: BBa_K812233
- And with a fluorescent citrine to characterize the promoter pElastase in the frog:
- We also submitted a heat shock Xenopus promoter: BBa_K812331
A serie of new eukaryotic reporters ready for expression
We also prepared a series of fluorescent reporters with a kozak sequence for high expression in eukaryotes.
And provided them directely cloned in pSC2+ for direct expression in Xenopus or chicken:
All these plasmids can be directly injected in to the Xenopus embryo after a miniprep, and are designed for rapid testing of multicellular devices. They contain all sequences necessary for a tissue specific expression, as well as debugging tools. Several plasmids can be co-injected, allowing ratio adjustment and linking the system do modeling.Once the system is tested and debugged, the final system can be implemented by chromosomal insertion.
Characterization
The information on characterized part can be found here: FrenchFrogThe auxin system
Auxin production device
We adapted the auxin production module created by the Imperial College 2011 team for it to be used in eukaryotes. We made a system designed for coinjection, and a system which works with a single “operon” using pep2A. This is a short self-cleaving peptide, which will cut a nascent peptide chain to make 2 proteins, insuring stochiometry of the products in the cell. These auxin production devices are designed to communicate through the organism with the auxin reception device.
- K812020: IAAH enzyme that catalyzes the second and last step towards auxin production
- K812021: IAAM enzyme that catalyses the transformation of Tryptophan to Inodle-3-acetamide
- K812014: Auxin production cassette containg both enzymes above which is monocystronic
Auxin reception device
The auxin reception device is composed of two components : Tir1, an auxin dependant ubiquitin ligase, and AID – a peptide that is recognized and ubiquitinated specifically by Tir1, when auxin is present. By fusing AID to any protein, this protein will be degraded along with the AID tag when auxin is added to the medium.
- K812010: GFP fusion with the ubuquitinase E3 OsTirI recoginition domain which a main part of the degron system
- K812012: OsTirI Ubiquitinase E3 for AID tagged protein degradation in the presence of auxin
- K812013: The Auxin reciever system golden gate-assembled containing GFP-AID OsTirI polysistronic system for auxin detection in tadpole
Goldenbricks
Summary
The GoldenBrick is a new assembly method for the partsregistry. If you want to know more, see this page.
Our favourites parts
- K812050: A GoldenBricked version of pSB1C3 with J04450 as negative cloning control
- K812051: A GoldenBricked version of pSB1K3 with J04450 as negative cloning control
The other parts we have created
- K812050: A GoldenBricked version of pSB1C3 with J04450 as negative cloning control
- K812051: A GoldenBricked version of pSB1K3 with J04450 as negative cloning control
- K812053: A GoldenBricked version of the strong RBS B0034
- K812054: A GoldenBricked version of the RFP E1010
- K812055: A GoldenBricked version of the terminator B0015
- K812056: A GoldenBricked version of the pLac R0010 promoter
- K812057: A GoldenBricked of an sfGFP protein
- K812058: A GoldenBricked of medium strenght RBS J61107
- K812059: A GoldenBricked of week RBS J61117
 "
"