Team:Cornell/project/wetlab/results/nah operon
From 2012.igem.org
(Difference between revisions)
R.Lizarralde (Talk | contribs) |
|||
| (15 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
</li> | </li> | ||
<li> | <li> | ||
| - | <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/ | + | <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/transcription">Transcriptional Characterization</a> |
</li> | </li> | ||
<li> | <li> | ||
| - | <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/ | + | <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/currentresponse">Current Response</a> |
| - | </li> | + | </li> |
| - | + | ||
<li> | <li> | ||
<a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/protein">MtrB Protein Expression</a> | <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/protein">MtrB Protein Expression</a> | ||
| Line 72: | Line 71: | ||
<div class="row"> | <div class="row"> | ||
<div class="nine columns"> | <div class="nine columns"> | ||
| - | <h3> | + | <h3>Biosynthesis of Indigo</h3> |
| - | + | Past work indicates that when the <i>nah</i> operon from the NAH7 <i>Pseudomonas</i> plasmid is heterologously expressed in <i>E. coli</i>, the cells have the ability to synthesize indigo from tryptophan [1]. Therefore, as indirect proof that our <i>nah operon</i> BioBrick has activity in our engineered strains, we set several cultures to determine whether indigo was present when cultures were supplemented tryptophan. <br><br><i>E. coli</i> contains the enzyme tryptophanase, which splits the indole group from the peptide backbone. Naphthalene dioxygenase, from the <i>nah</i> operon, then creates a compound which will spontaneously eliminate water and be air oxidized into indigo as the final product. | |
| + | <br><br>We are in the process of isolating the four coding regions which make up the naphthalene dioxygenase complex from the operon and <a href="https://2012.igem.org/Team:Cornell/project/wetlab/results/biobricks">BioBrick-ing</a> this genetic part so that future researchers have access an indigo-producing BioBrick. | ||
</div> | </div> | ||
<div class="three columns"> | <div class="three columns"> | ||
| - | <img src="https://static.igem.org/mediawiki/ | + | <a href="https://static.igem.org/mediawiki/igem.org/8/88/IMG_0149notswqua.jpg" rel="lightbox"><img src="https://static.igem.org/mediawiki/igem.org/e/e3/IMG_0149_square.jpg"></a> |
</div> | </div> | ||
</div> | </div> | ||
| - | <div class="row | + | <div class="row"> |
<div class="three columns"> | <div class="three columns"> | ||
| - | <img src="https://static.igem.org/mediawiki/ | + | <a href="https://static.igem.org/mediawiki/igem.org/a/aa/IMG_0133not.jpg" rel="lightbox"><img src="https://static.igem.org/mediawiki/igem.org/9/95/IMG_0133square.jpg"></a> |
</div> | </div> | ||
<div class="nine columns"> | <div class="nine columns"> | ||
| - | <h3> | + | <h3>Testing and Results</h3> |
| - | + | Cultures of DH5α containing the <i>nah</i> operon were started in LB broth with 200 μM tryptophan and 100 mg/L ampicillin. Six days after the initial cultures were started, the liquid culture appeared purple. After extracting with chloroform, the organic phase was a light purple color. Testing is being done to determine if this purple compound is indigo. | |
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 97: | Line 94: | ||
</div> | </div> | ||
| + | <div class="twelve columns"> | ||
| + | <h3>References</h3> | ||
| + | [1] Ensley, B., Ratzkin, B., Osslund, T., Simon, M., Wackett, L., & Gibson, D. (1983). Expression of naphthalene oxidation genes in <i>Escherichia coli</i> results in the biosynthesis of indigo. <i>Science</i>, 222(4620), 167-9. | ||
| + | <br></br> | ||
| + | |||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 02:38, 27 October 2012
nah Operon Expression
Biosynthesis of Indigo
Past work indicates that when the nah operon from the NAH7 Pseudomonas plasmid is heterologously expressed in E. coli, the cells have the ability to synthesize indigo from tryptophan [1]. Therefore, as indirect proof that our nah operon BioBrick has activity in our engineered strains, we set several cultures to determine whether indigo was present when cultures were supplemented tryptophan.E. coli contains the enzyme tryptophanase, which splits the indole group from the peptide backbone. Naphthalene dioxygenase, from the nah operon, then creates a compound which will spontaneously eliminate water and be air oxidized into indigo as the final product.
We are in the process of isolating the four coding regions which make up the naphthalene dioxygenase complex from the operon and BioBrick-ing this genetic part so that future researchers have access an indigo-producing BioBrick.
Testing and Results
Cultures of DH5α containing the nah operon were started in LB broth with 200 μM tryptophan and 100 mg/L ampicillin. Six days after the initial cultures were started, the liquid culture appeared purple. After extracting with chloroform, the organic phase was a light purple color. Testing is being done to determine if this purple compound is indigo.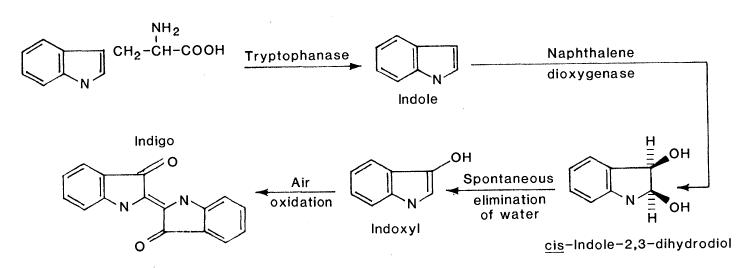 Fig. 1. Pathway by which E. coli can produce indigo from tryptophan when expressing genes of the Nah7 operon. (Ensley et al., 1983)
Fig. 1. Pathway by which E. coli can produce indigo from tryptophan when expressing genes of the Nah7 operon. (Ensley et al., 1983)
 "
"









