From 2012.igem.org
The purpose of this subproject is to make the nah operon biobrick compatible for the use of future iGEM teams. The nah operon codes for a pathway that converts naphthalene into salicylate. In our system, this will allow naphthalene to be detected by the salicylate reporter. There are three standard cut sites internal to the operon which we are removing - PstI, and two NotI sites. This biobrick compatible piece could be useful to future teams in projects that involve bioremediation or detection of naphthalene.
June
July
June 17th-23rd
June 17th, Sunday
June 19th, Tuesday
- Ran Gibson Assembly of nah operon fragments into pBMT-1 backbone and transformed into DH5a electrocompotent E. coli cells.
- Set up PCR to amplify the Gibson Assembly products.
- Work done by: Dylan and Swati
June 20th, Wednesday
- Set up a digestion of the PCR of Gibson Assembly products and ran the digested products on a gel to see if Gibson worked.
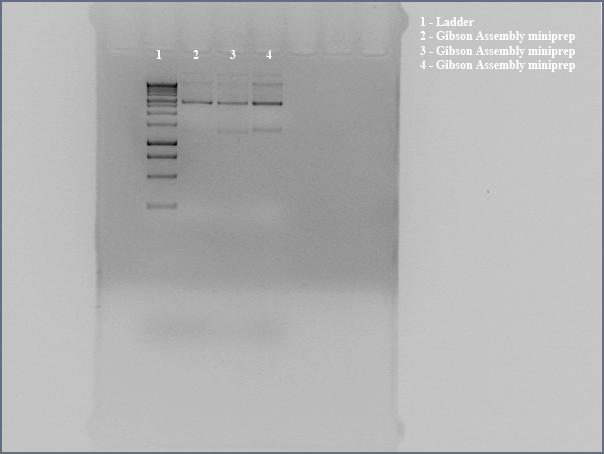
- Three colonies on plates of DH5a transformed with Gibson Assembly product. Made liquid cultures to miniprep and sequence.
- Work done by: Dylan and Swati
June 22nd, Friday
- Miniprepped directly transformed Gibson Assembly product for sequencing using the the Gibson nah1F and nah4R primers (each w/ 20 bp overhangs).
- Ran undigested miniprep with gel electrophoresis, looking for large bands corresponding to supercoiled DNA. The gel shows distinct bands for all three lanes. We interpreted this to mean that we got product. Submitted for sequencing
June 24th-30th
June 27th, Wednesday
Rafael and Dylan making an agarose gel.
====June 28th, Thursday====
 "
"

