Team:Cornell/Dylan Scratch Notebook
From 2012.igem.org
| Home | Team | Project | Parts | Modeling | Notebook | Protocols | Safety | Attributions |
|---|
June
June 12th-16th
June 12th, Tuesday
Today, we began bootcamp!Click to view the details.
These are more details. These are more details. These are more details. These are more details. These are more details. These are more details. These are more details. These are more details. These are more details. These are more details.
June 13th, Wednesday
Click to view the details.
- Set up PCR for four fragments of the nah operon as well as the pBMT-1 backbone, using primers designed to mutate cut-sites concurrently with Gibson Assembly. Also set up a PCR for the entire nah operon. Due to length of the fragments, a longer extension time was chosen.
- If Gibson Assembly fails, but we are able to PCR the entire 10kb nah operon, an alternate method of mutation using three sequential site-directed mutageneses will be pursued.
- Work done by: Caleb, Claire, Dylan, Spencer, Steven, and Swati
June 14th, Thursday
Click to view the details.
PCR only amplified nah1, nah3, and nah4 - longer fragments (nah2, the full nah operon, and pBMT-1) were not identified on the PCR product gel. Set up another PCR with shorter extension time.
June 15th, Friday
Click to view the details.
PCR of four out of five products visible on gel. Set up PCR of pBMT-1, final fragment required for Gibson Assembly of the nah operon.
June 16th, Saturday
Click to view the details.
June 17th-23rd
June 17th, Sunday
- Successful PCR of pBMT-1 gel purified.
June 18th, Monday
June 19th, Tuesday
- Ran Gibson Assembly of nah operon fragments into pBMT-1 backbone and transformed into DH5α electrocompotent E. coli cells.
- Set up PCR to amplify the Gibson Assembly products.
- Work done by: Dylan and Swati
June 20th, Wednesday
- Set up a digestion of the PCR of Gibson Assembly products and ran the digested products on a gel to see if Gibson worked.
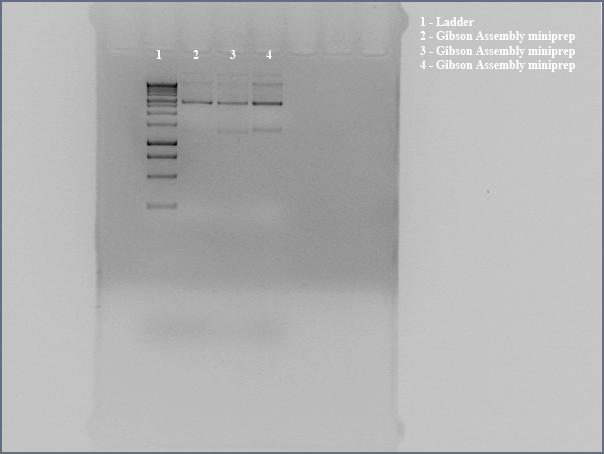
- Three colonies on plates of DH5a transformed with Gibson Assembly product. Made liquid cultures to miniprep and sequence.
- Work done by: Dylan and Swati
June 22nd, Friday
- Miniprepped directly transformed Gibson Assembly product for sequencing using the the Gibson nah1F and nah4R primers (each w/ 20 bp overhangs).
- Ran undigested miniprep with gel electrophoresis, looking for large bands corresponding to supercoiled DNA. The gel shows distinct bands for all three lanes. We interpreted this to mean that we got product. Submitted for sequencing
June 23rd, Saturday
June 24th-30th
June 24th, Sunday
June 25th, Monday
June 26th, Tuesday
June 27th, Wednesday
- Ran digest of gibson-assembled nah operon with gel electrophoresis
June 28th, Thursday
- Gel purified arsR construct
June 29th, Friday
- Vent PCR at 11:00 (DPW)
- Amplifying both previous Phusion PCR band and original p21 template
- Dylan's magic triple anneal method (55/60/63)
- Gel purified PCR product from Phusion template (~1:20 pm) (DPW)
- Quantified product at 22.4 ng/uL
- Set up digestion of p21 PCR product with EcoRI-HF and AscI (~9:00 pm).
- 22 ng/uL --> 45.5 uL sample for 1 ug digest
- Buffer 4
- Ran digestion on gel. (~11:00 pm)
- Sliced out relevant band on gel, stored overnight at -20.
- Miniprepped overnight cultures of PL14-PL20 (~1:00 pm, STC)
- Using C1015 rotor, 6666 x g, the Corning culture tubes only fit in the centrifuge with the lids off
- Made LB, 3x 60 mL in 100 mL bottles (~ 3:00pm, SS)
- Made LB Agar, 4 x 250 mL LB Agar in 500 mL bottles (~3:00, CS)
- Autoclaved LB, LB Agar, and milliQ (~3:30 pm, SS)
- Made LB plates with Kan (~6:30 pm, CMR)
- CUGEM movie outing at 8:00 pm.
- Phusion PCR at 10:00 pm (DPW)
- Dylan's magic triple anneal method (57/65/70, 35 cycles total)
- Amplifying nah operon from Gibson 1
- Appending BioBrick cutsites for ligation into pSB3C5
June 30th, Saturday
- Took PCR out of thermal cycler at 9:00 am (DPW)
- Set up gel using NEB 100bp and 2-log ladders (10:00am)
- Gel extracted PCR product, quantified at ~10ng/uL
- Set up new Phusion PCR using Gibson 1 as template
- Dylan's magic three-anneal method (57.6/65/72)
- Extension time of 3 min.
- Continued gel extraction of p21 PCR digest from previous day (SS)
- Set up ligation of p21 PCR digest extraction and dephosphorylated pBBRBB+mtrB (11:11am, SS)
- Desalted ligation using Millipore membrane paper
- Transformed 2 electrocompetent DH5alpha stocks at 5:30 pm and 5:50 pm, respectively.
- Observed time constants for electroporation of 4.38 ms and 4.24 ms, respectively
- Let cells recover for 1 hour, plated on LB + Kan.
- Set up two ligations of pSB3C5 into PNNL electrocompetent Shewanella strain JG700 (6:30 pm, Sp.C and St.C)
- First transformation performed at usual PNNL voltage of 0.75 kV (time constant of 9.30 ms)
- Second transformation performed at Myers and Myers specification of 0.55 kV (time constant of 9.34 ms).
 "
"