Team:Chalmers-Gothenburg/Methods
From 2012.igem.org
Contents |
Methods
The work required for the construction of the biosensor is divided into three main tasks: (1) expression of the LH/CG receptor, (2) knock-out of the CWP2 gene encoding a cell wall mannoprotein and (3) expression of indigo synthesizing enzymes tryptophanase and monooxygenase. These tasks will be performed in parallel and in the end, the different parts will be combined into one system. The three main tasks are described in detail in the following sections.
Expression of the LHCGR gene in yeast strain IMFD-73
The first task is to express the LH/CG receptor in yeast. The receptor contains several amino acid residues that are essential for efficient ligand binding, both in the extracellular and in the transmembrane domains. In addition, the C-terminus and the intracellular loop III are considered to bind to the associated Gα protein of the receptor, and thereby constitute an important part of the signal transduction [1]. In order to ensure correct conformational change following ligand binding and to guarantee a proper signal transduction, the full-length LH/CG receptor will be expressed. The LHCGR gene will be inserted into the yeast strain IMFD-73 and be placed under the control of a strong, constitutive promoter to ensure constant expression. To enhance translation efficiency, a Kozak sequence (AAAACA) will also be inserted upstream of the LHCGR gene. In addition, a signal peptide will be added to the N-terminus of the receptor sequence to ensure transportation to the yeast cell membrane.
Previous research does not provide general guidelines for which signal peptide should be used when expressing human GPCRs in yeast. In this project, both the native signal of the LH/CG receptor, consisting of its 22 first amino acids [2], and a yeast signal peptide will be tested separately. The 5' end of the LH/CG receptor, which will be used as the native signal peptide, is originally glycosylated and glycosylation in yeast differs significantly from the one in human [3]. The native signal will, however, be tested even though it is not sure if glycosylation of these amino acid residues is required for correct transportation of the receptor to the membrane [4]. In order to test a yeast signal, the idea was to fuse the signal peptide of Ste2, the receptor to be replaced, with the LH/CG receptor N-terminus. However, Ste2 lacks an identified signal sequence [5]. Instead, the signal peptide from Ste3, the MATα type pheromone receptor, will be used. It consists of the first 25 amino acid residues of the receptor [5].
Plasmid construction
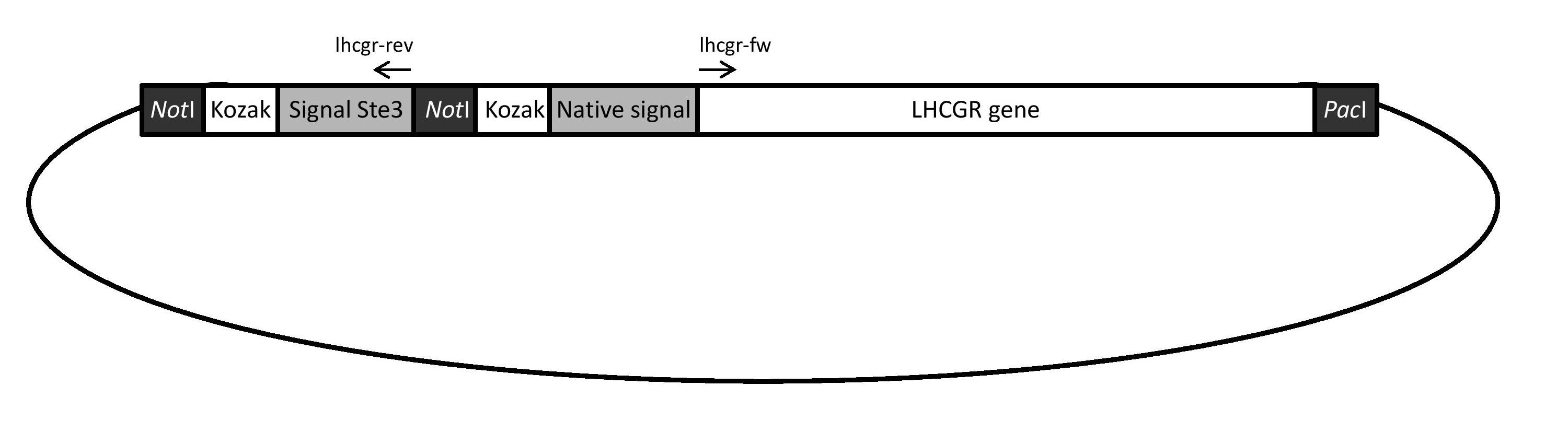
The LHCGR gene construct will be ordered in a vector. Figure 1 provides a schematic illustration of this construct. The signal peptide of Ste3 and a Kozak sequence will be fused with a Kozak sequence at the 5' site of the native signal and the gene. In addition, NotI and one PacI restriction sites will be added as indicated in Figure 1.
Figure 1: Schematic illustration of the LHCGR gene construct. Using this construct enables expression of the LHCGR gene with each signal peptides separately. For the expression of the gene with the native signal, the vector can be digested directly with NotI and PacI. When testing the expression with the yeast signal, PCR can be run with 5' phosphorylated primers indicated by arrows in order to exclude the native signal and its associated Kozak sequence and PacI restriction site. The resulting linear fragment can be religated.
The aim of using this particular construct is to enable expression of the LHCGR gene with both signal peptides separately. For the expression with the native signal, the vector containing the LHCGR gene will be digested directly with NotI and PacI restriction enzymes. The fragments will be purified on a gel and then ligated into the corresponding sites of the expression plasmid pSP-GM1 with bidirectional promoters TEF1 and PGK1, a URA3 marker gene and an ampicillin resistance gene. For expression of the LHCGR with the Ste3 signal peptide, PCR will be run with 5' phosphorylated primers indicated with arrows in Figure 1 (lhcgr-rev and lhcgr-fw). This will result in a linear fragment that lacks the native signal and its associated NotI site and Kozak sequence. This fragment will be purified on a gel, religated and amplified in E. coli. After that, it will be digested with NotI and PacI restriction enzymes, column purified and then ligated into the expression plasmid pSP-GM1.
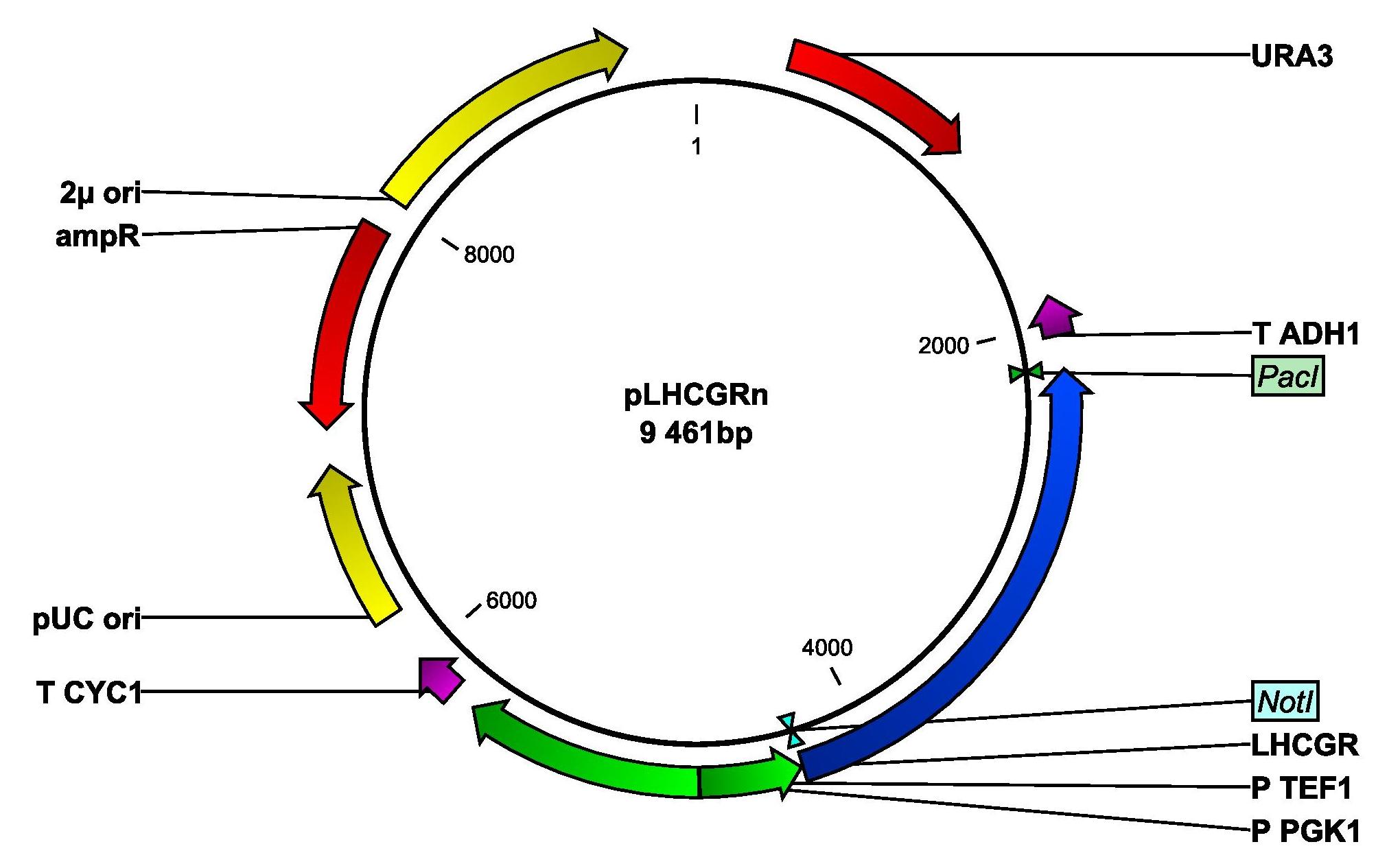
In both cases, the gene will be set under the control of the strong and constitutive TEF1 promoter. The resulting plasmids will be named pLHCGRn (native signal peptide) and pLHCGRy (yeast signal peptide). Figure 2 shows a map of the plasmid pLHCGRn. pLHCGRy is similar to pLHCGRn and only differs in length of the LHCGR gene construct, which is 9 bp longer in the pLHCGRy plasmid. For amplification, E. coli strain DH5α will be transformed separately with the two plasmids and an empty control plasmid by heatshocking. The cells will be cultivated in ampicillin-containing LB medium.
Figure 2: Plasmid map of pLHCGRn derived from pSP-GM1. The plasmid contains a yeast origin of replication, 2μ ori, and a yeast marker, URA3. The parts that are important for transforming E. coli are the ampicillin resistance gene, AmpR and the E. coli origin of replication, pUC ori. The LHCGR gene construct with the yeast signal peptide is anked by NotI and PacI. P TEF1 is a strong, constitutive promoter and T ADH1 is a terminator.
Transformation of IMFD-73 and degradation of the cell wall
After amplification in DH5α, the plasmids will be purified from E. coli. The yeast strain IMFD-73 will be transformed with either pLHCGRn or pLHCGRy using the High Efficiency Transformation (TRAFO) method. As a control, a third transformation of IMFD-73 with the empty expression plasmid will also be performed. The cells will be cultivated and selected in uracil-deficient media. After transformation, hCG hormone, ordered from a company, will be added and screening for fluorescence will be performed. As mentioned previously, proteins consisting of 70 or more amino acids have very limited possibilities of penetrating the yeast cell wall [6]. Since the hCG hormone has two subunits consisting of 92 and 145 amino acid residues, respectively, it is likely that it will not be able to pass the cell wall. Therefore, Zymolyase, a cell wall degrading enzyme, will be added and the screening procedure will be repeated. For further experiments, the plasmid (either pLHCGRn or pLHCGRy) expressing the more functional receptor will be chosen. The fluorescence levels will therefore be compared and the plasmid of the strain displaying the higher fluorescence intensity will be used. The strain with the empty control plasmid will be used to examine the background fluorescence signal and serves as a reference.
Deletion of CWP2
The second task focuses on modifying the IMFD-73 strain in such a way that its cell wall is weakened. The aim is for the hCG hormone to be able to pass the cell wall without the need of adding Zymolyase, which is impractical if the aim for the test kit to be used at home. Furthermore, removing the cell wall with enzymes results in less stable cells and this is not desirable if the construction should be used as a biosensor. Hence, instead of removing the whole cell wall, the IMFD-73 strain will have the gene CWP2 deleted in order to decimate the cell wall. This gene encodes a mannoprotein that is a major constituent of the cell wall and important for its stabilization [5]. Previously, it has been reported that CWP mutants have an increased cell wall permeability [7] which is a desired feature of the hCG biosensor.
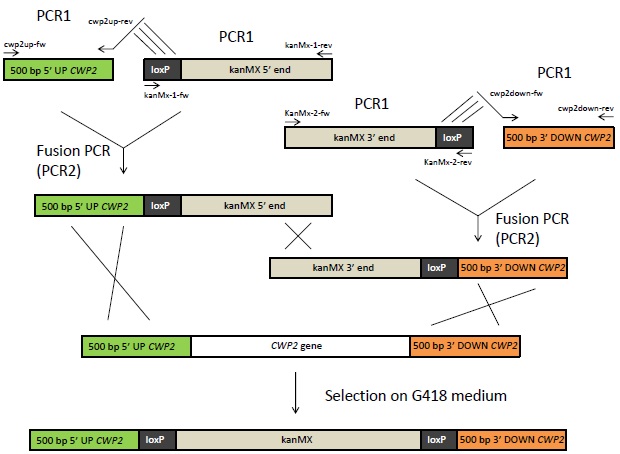
The deletion of CWP2 will be done using the Bipartite method, which is outlined in Figure 3. For this, four DNA fragments will be needed. Two of those will consist of 500 bp from the upstream respectively downstream region of the CWP2 gene. The other two fragments will consist of the 5' respectively 3' end of the kanMX cassette. This cassette contains a kanamycin resistance gene under control of the TEF1 promoter and terminator. Each of these two fragments will also contain a loxP site. All four fragments will first be amplified in separate PCR reactions (denoted “PCR1” reactions in Figure 3). The kanMX fragments, which will be available in already existing plasmids, are amplified using the primers kanMx-1-fw, kanMx-1-rev, kanMx-2-fw and kanMx-2-rev. The CWP2 fragments will need to be obtained and amplified from yeast via colony-PCR using primers cwp2up-fw, cwp2up-rev, cw2pdown-fw and cwp2down-rev. The primers cwp2up-rev and cwp2down-fw have 5' extensions that are reverse complement (indicated with lines in Figure 3) to the loxP sites of the kanMX fragments. The PCR1 reaction with the CWP2 up and downstream sequences will consequently generate two fragments that are overlapping the loxP sites in the kanMX fragments.
The next step will be to fuse the CWP2 up- and downstream region to the kanMX gene by fusion PCR as indicated in Figure 3. In this second PCR reaction, the CWP2 upstream fragment that resulted from PCR1 and the overlapping kanMX 5' end fragment will together serve as templates. PCR will be run with primers cwp2up-fw and kanMx-1-rev. The same principle will be applied in order to fuse the kanMX 3' end with the CWP2 downstream fragment. The resulting two fragments of PCR2 will then be purified and inserted into IMFD-73, using the TRAFO method. Through homologous recombination, the CWP2 gene will then be replaced by the kanMX cassette. The cells will be selected on G418 medium, where only cells containing the kanMX gene will be able to grow. The deletion will also be verified by verification PCR. The loxP site can be used to remove the marker after selection, but this will not be necessary in this case.
Figure 3: Overview of the Bipartite method. 500 bp of the up- and downstream sequence of the CWP2 gene will be amplified from genomic DNA using primers with 5' extensions that are complement to the loxP sites as indicated with lines above. The 5' and 3' end of the kanMX (kanamycin resistance) gene flanked to loxP sites, available in vectors, will also be amplified by PCR. The fragments from these PCR1 reactions will be fused by Fusion PCR. The resulting fragments of this reaction can then be inserted into yeast. Homologous recombination will lead to the exchange of CWP2 with kanMX in the yeast's genome. The cells can be selected on G418 medium.
Expression of monooxygenase and tryptophanase genes
The third task in this project is to manipulate the yeast cell in order to produce indigo. The dye indigo is chosen as the output signal in the pregnancy test due to its high intensity. A small amount of this molecule will produce a powerful stain. Two enzymes are needed for the synthesis of indigo from the precursor tryptophan: tryptophanase and a mono- or a dioxygenase. Expression of the fmo gene encoding the avin-containing monooxigenase in E. coli has resulted in the most successful bio-indigo production [8]. Therefore, the fmo gene will be used in this project. A gene with similarities to fmo has been identified in yeast [9] but due to lack of available research records, the fmo gene from Methylophaga will be used instead. The tryptophanase gene (tnaA) will be cloned from E. coli. Both genes will then be inserted into S. cerevisiae and will at first be set under the control of constitutive promoters. This is done in order to test if it is possible to produce indigo in yeast. If bio-indigo production is successful, the constitutive promoter for the tnaA gene will be exchanged by the FIG1 promoter in order to couple the bio-indigo production with the pheromone pathway. The plasmid can then be used to transform IMFD-73.
Plasmid construction and transformation of a yeast lab strain
Two cloning steps are needed to construct the plasmid with the tnaA and fmo genes. E. coli strains contain the gene coding for tryptophanase (tnaA) and the sequence will be amplified from genomic DNA using colony-PCR. The used primers are tnaa-fw and tnaa-rev. To the 5' end of the primers, BamHI and XhoI sequences are added in order to flank the gene sequence with these restriction sites. The PCR product will be purified on gel and digested with BamHI and XhoI. After digestion, the fragment will be purified on a column and ligated into the corresponding restriction sites of the expression plasmid pIYC04 with the bidirectional, constitutive promoters TEF1 and PGK1, a HIS3 marker gene and an ampicillin resistance gene. The tnaA gene will be set under the control of the PGK1 promoter. For amplification of the plasmid, E. coli strain DH5α will be transformed by heatshocking and the cells will be cultivated in ampicillin-containing media. Afterwards, the plasmid will be purified.
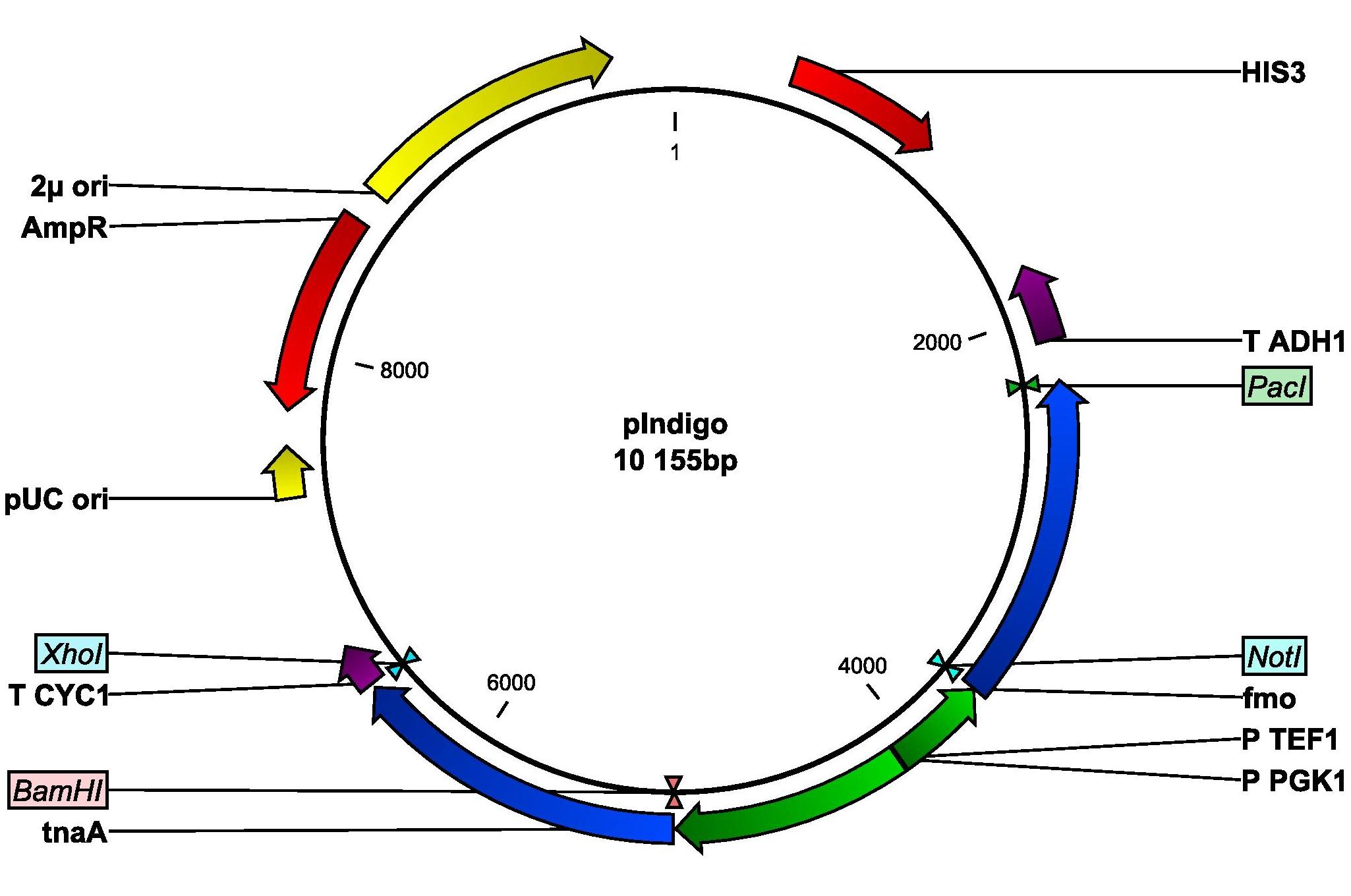
The gene for the monooxygenase (fmo), derived from Methylophaga, is kindly provided by Si Wouk Kim from Chosun University in South Korea. The sequence will be amplified by PCR with primers fmo-fw and fmo-rev. Restriction sites NotI and PacI are inserted into the primers. The PCR product will be purified on gel and digested with restriction enzymes NotI and PacI. The digested fragments will then be purified on a column and ligated into the expression plasmid containing the tnaA gene. Thus, the fmo gene is set under the control of the TEF1 promoter. The resulting plasmid will be called pIndigo and amplified in E. coli. A plasmid map of pIndigo is shown in Figure 4. After amplification, the plasmid will be used to transform the normal yeast lab strain CEN.PK 113-11C using the TRAFO method. If the experiments are successful, indigo will be produced. If indigo is not detected, it might be due to low intracellular levels of tryptophan, which will therefore be added to the culture and screening for indigo will be repeated.
Figure 4: Plasmid map of pIndigo derived from pIYC04. The plasmid contains a yeast origin of replication, 2μ ori and a yeast marker, HIS3. The important parts for transforming E. coli are the ampicillin resistance gene, AmpR, and the E. coli origin of replication, pUC ori. The fmo gene is flanked by NotI and PacI and is set under the control of the TEF1 promoter. The tnaA gene is flanked by BamHI and XhoI and is set under the control of the PGK1 promoter. T CYC1 and T ADH1 are terminators.
Replacement of the PGK1 with the FIG1 promoter
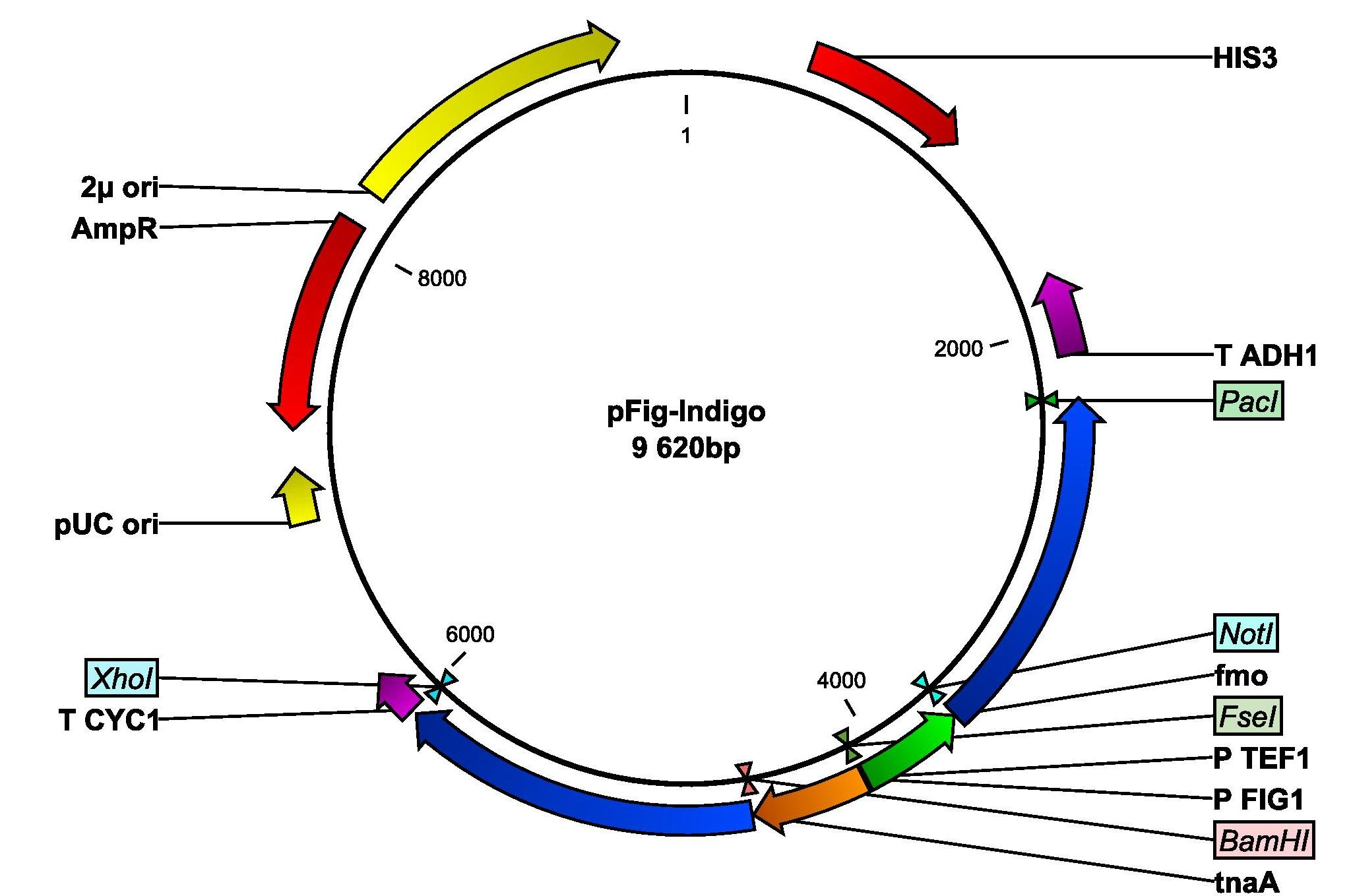
The next step is to replace the PGK1 with the FIG1 promoter. 450 bp of the 5' upstream regulatory sequence of the FIG1 gene will be amplified from genomic DNA from a normal yeast strain using colony-PCR. The primers fig-fw and fig-rev will used. Into the primers, BamHI and FseI restriction sites are inserted. The PCR product will be purified on gel, digested with the restriction enzymes and ligated into the pIndigo, hence removing PPGK1 and inserting P(FIG1). This plasmid will be named pFig-Indigo and is illustrated in Figure 5.
Figure 5: Plasmid map of pFig-Indigo derived from pIndigo. The plasmid contains a yeast origin of replication, 2μ ori and a yeast marker, HIS3. The important parts for transforming E. coli are the ampicillin resistance gene, AmpR, and the E. coli origin of replication, pUC ori. The fmo gene is flanked by NotI and PacI and is set under the control of TEF1 promoter. The tnaA gene is now set under the control of the FIG1 promoter. T CYC1 and T ADH1 are terminators.
Combination of the components to a functional biosensor
The final step of the project will be to combine the parts described above in one single host strain. The IMFD-73 with the deleted CWP2 gene from task 2 will be transformed with either pLHCGRy or pLHCGRn using TRAFO and the cells will be selected on uracil-deficient media. After that, the pFig Indigo will also be inserted using TRAFO and the cells will be grown on histidine-deficient media. The ability of the resulting yeast to function as a biosensor will then be tested by adding different concentrations of the hCG hormone and screening for output signals will be done.
References
[1] Ryu KS, Ji L, Chang L, Ji TH. Molecular mechanism of LH/CG-receptor activation. Molecular and Cellular Endocrinology. 1996;125(1-2):93-100. [2] Dufau ML. The luteininizing hormone receptor. Annual review of Physiology. 1998;60(1):461-496. [3] Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Current Opinion in Biotechnology. 2007;18(5):387-392. [4] Petäja-Repo UE, Merz WE, Rajaniemi HJ. Significance of the carbohydrate moiety of the rat ovarian luteinizing-hormone/chorionic-gonadotropin receptor for ligand-binding specificity and signal transduction. The Biochemical Journal. 1993;292(3):839-844. [5] Saccharomyces Genome Database [Online]. Stanford: National Human Genome Research Institute; 1997 [updated 2012 Feb 9; cited 2012 May 13]. Available from: http://www.yeastgenome.org/. [6] Ladds G, Goddard A, Davey J. Functional analysis of heterologous GPCR signaling pathways in yeast. Trends in Biotechnology. 2005;23(7):367-373. [7] Zhang M, Liang Y, Zhang X, Xu Y, Dai H, Xiao W. Deletion of yeast CWP genes enhances cell permeability to genotoxic agents. Toxicological sciences : an official journal of the Society of Toxicology. 2008;130(1):68-76. [8] Han GH, Shin HJ, Kim SW. Optimization of bio-indigoproduction by recombinant E. coli harboring fmo gene. Enzyme and Microbial Technology. 2008;42(7):617-623. [9] Choi HS, Kim JK, Cho HE, Kim YC, Kim JI, Kim SW. A novel flavin-containing monooxygenase from Methylophaga sp.strain SK1 and its indigo synthesis in Escherichia coli. Biochemical and Biophysical Research Communications. 2003;306(4):930-936.
 "
"