Team:Cambridge/Lab book/Week 8
From 2012.igem.org
(→Saturday (18/08/12)) |
|||
| Line 199: | Line 199: | ||
*Results: failed- no wanted bands, since the positive control didn't come up, it is suggested that we need to optimise the PCR protocol for Velocity if we are to use it. | *Results: failed- no wanted bands, since the positive control didn't come up, it is suggested that we need to optimise the PCR protocol for Velocity if we are to use it. | ||
| + | |||
| + | |||
| + | ===Sunday (19/08/12)=== | ||
| + | |||
| + | '''Ratiometrica-Flu: [[Team:Cambridge/Protocols/PCRProtocol|3rd attempt at PCR of fragments]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Similar PCR to the one from yesterday, except Velocity mastermix is further changed, and extension time is changed | ||
| + | |||
| + | *Results: failed- no bands, probably Velocity mastermix problem, will revert back to Phusion on next attempt | ||
{{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | ||
Revision as of 01:42, 27 September 2012
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|
Contents |
Monday (13/08/12)
Ratiometrica-Lux: Miniprep of LuxBrick plasmid
- Miniprepped is done using miniprep kit supplied by Cambio
- Plasmids are miniprepped from 3 cultures
Ratiometrica-Lux: PCR of lux vector
- With split primers, similar conditions to last week (except no longer using E. coli colonies), each half in triplicates
- Results: Half of the vector (Fragment B) came out (gel 1), positive control worked (gel 2 lane 5)
Ratiometrica-Lux: Arabinose induction of Lux E. coli
- E. coli with the lux plasmid (K325909) O/N liquid cultures are induced with 3mM arabinose
- They did not produce visible bioluminescence after 7 hours. We suspect it to be a problem associated either with the concentration of cells in the culture or the amount of arabinose we added.
Tuesday (14/08/12)
Gibson assembly of positive control
- Fragments from Tom's PCR from last week are used: the PSB4K5 backbone (in triplicates), and sfGFP from sfGFP-ampR (in triplicates)
- Protocol changed slightly: 0.5 μl of each DNA fragment solution mixed in with master mix to make up 4 μl total volume (1μl DNA and 3μl master mix).
Transformation of e.coli with positive control DNA
- Chemically competent e.coli cells transformed with plasmid DNA produced by Gibson assembly step.
- Transformants plated out on 50 μg/ml kanomycin plates. Incubated overnight at 37 °C.
Ratiometrica-Lux: Arabinose Induction of Lux E. coli
- After our failed liquid culture induction, we made LB agar plates with 3mM arabinose and the appropriate antibiotics.
- Cells are plated out on the arabinose plates and incubated at 37°C overnight.
- Colonies on plates show bioluminescence after overnight incubation
Wednesday (15/08/12)
PCR of positive control fragments
- A replication of last week's PCR,except each fragment is done 5 times to generate more fragments so that the positive control could be used for future experiments
- Results: successful amplification of all fragments.
Extraction of positive control DNA
- Positive control DNA from gel excised and purified.
- Additional elution steps used to concentrate resultant solution: eluants are recycled at least once into the spin column
- Verified with nanodropper - final DNA concentrations are around 20ng/ul for first eluants, and 15ng/ul for second eluants; this is twice as concentrated as before
Gibson assembly of positive control DNA
- A combination of fragments from the PCR are chosen: the highest, second highest, third highest, lowest concentration first eluants, and the highest concentration second eluants are put together
- Reading that our T5 exonuclease concentration in Gibson assembly may be too high, we made up some mastermix with 1/5 of the T5 exo concentration and tested that in parallel with the fragments of various concentrations (2x5 = 10 Gibson reactions in total)
- Results: Successful Gibson assembly reactions. The lowered T5 exo conc seems to lower the Gibson efficiency, and the more concentrated fragments seem to produce more colonies.
Transformation of positive control DNA into e.coli
- Chemically competent e.coli transformed with positive control Gibson products made previously.
- Transformants plated out onto 50 μg/ml kanomycin plates and incubated at 37 °C overnight.
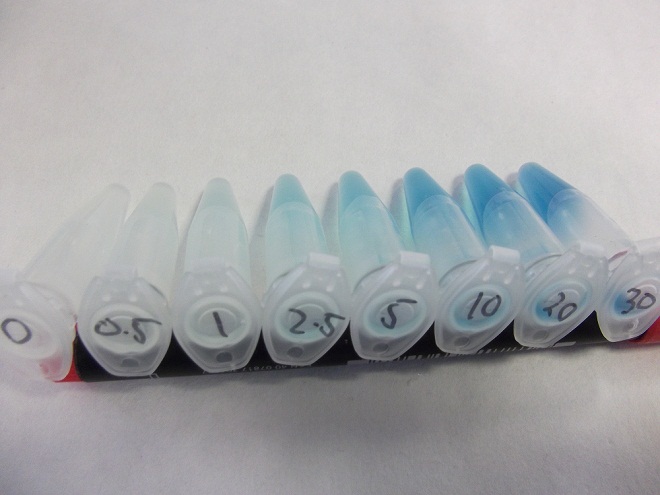
Characterization of fluoride riboswitch construct
- Strain of bacillus lacking fluoride transporter system tested. Concentrations used:
- 0mM - 0.5mM - 1mM - 2.5mM - 5mM - 10mM - 20mM - 30mM
Thursday (16/08/12)
Characterization of fluoride riboswitch construct
- Eppindorfs containing the bacillus with the fluoride riboswitch removed from incubator and imaged.
- Now this definitely works, we will try quantifying this riboswitch with an ONPG assay and the plate reader.
PCR ran by paul - insert photos/discuss
- Only positive control and two sets of triplicates worked
- YFP and CFP extracted successfully
Ratiometrica-Lux: Restriction Digest of Lux vector
- Results: Digest came out exactly as expected.
Ratiometrica-Flu: PCR of Flourescent Construct DNA fragments
- Redo of PCR done at the end of July, as we are running out of fragments to Gibson
- Used existing DNA fragments as template rather than biobricks from the registry
- Results: Only the fluorescent protein fragments (eCFP and eYFP) came out clearly; the smaller fragments (B0015, K143053) might have came out but was mistaken for primer dimers, the vector fragments did not come out
Friday (17/08/12)
- Team meet up today! No lab work was done.
Saturday (18/08/12)
RiboSense: PCR of Fluoride biobrick format
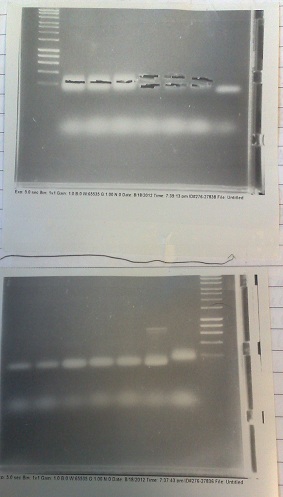
- Two sets of primers used: one to allow insertion into backbone by ligation, and one to allow insertion by Gibson assembly.
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. Fragments produced of correct size, however they overlapped with the primer dimer band due to the small size of the fragments.
- DNA extracted and purified.
Ribosense: PCR of Mg2+ riboswitch construct vector
- Separate reactions for
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. No fragments produced. Positive control worked however, so master mix clearly works.
- Given this PCR has worked in the past, will try to re-run with the same settings and hope for success in the future.
Ratiometrica-Flu: 2nd attempt at PCR of fragments
- Reattempt of pJS130 and smaller fragments
- Used Velocity from Bioline, new mastermix recipe is used
- Results: failed- no wanted bands, since the positive control didn't come up, it is suggested that we need to optimise the PCR protocol for Velocity if we are to use it.
Sunday (19/08/12)
Ratiometrica-Flu: 3rd attempt at PCR of fragments
- Similar PCR to the one from yesterday, except Velocity mastermix is further changed, and extension time is changed
- Results: failed- no bands, probably Velocity mastermix problem, will revert back to Phusion on next attempt
 "
"