Team:Calgary/Project/OSCAR/Decarboxylation
From 2012.igem.org
| Line 35: | Line 35: | ||
<h2>The PetroBrick</h2> | <h2>The PetroBrick</h2> | ||
| - | <p>The 2011 Washington iGEM team developed the PetroBrick, a BioBrick consisting of two primary genes. These include acyl-ACP reductase (AAR), which reduces fatty acids bound to ACP to fatty aldehydes, and a second gene called aldehyde decarbonylase (ADC), which subsequently cleaves the entire aldehyde group and results in a hydrocarbon chain. Essentially this allows for hydrocarbons to be produced from glucose. What we realized though, is that | + | <p>The 2011 Washington iGEM team developed the PetroBrick, a |
| + | |||
| + | BioBrick consisting of two primary genes. These include acyl-ACP | ||
| + | |||
| + | reductase (AAR), which reduces fatty acids bound to ACP to fatty | ||
| + | |||
| + | aldehydes, and a second gene called aldehyde decarbonylase (ADC), | ||
| + | |||
| + | which subsequently cleaves the entire aldehyde group and results | ||
| + | |||
| + | in a hydrocarbon chain. Essentially this allows for hydrocarbons | ||
| + | |||
| + | to be produced from glucose. What we realized though, is that | ||
| + | |||
| + | the fatty acids that the PetroBrick targets, have a very | ||
| + | |||
| + | similar structure to naphthenic acids.</p> | ||
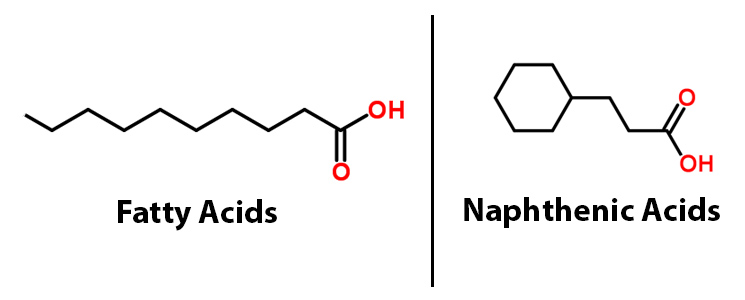
</html>[[File:UCalgary-Fatty-Acids-vs-NAs.jpg|500px|centre|thumb|Figure 1. A comparison of the structure of fatty acids and naphthenic acid]]<html> | </html>[[File:UCalgary-Fatty-Acids-vs-NAs.jpg|500px|centre|thumb|Figure 1. A comparison of the structure of fatty acids and naphthenic acid]]<html> | ||
| Line 41: | Line 57: | ||
| - | <p>This | + | <p>This lead us to believe that the PetroBrick may have the |
| - | + | ||
| + | potential to turn naphthenic acids in to hydrocarbons and | ||
| + | |||
| + | be a perfect solution to remediating naphthenic acids! First | ||
| + | |||
| + | though, we needed to show that the PetroBrick did in fact work as | ||
| + | |||
| + | expected. We had some difficulty with the DNA from the registry | ||
| + | |||
| + | and had to request the constructs directly from the Washington team. Once we had the Petrobrick, we needed to verify that the Petrobrick would work in our hands as it did for the 2011 Washington team. | ||
| + | |||
| + | Figures 2 and 3 demonstrate the function of the Petrobrick.</p> | ||
</html>[[File:Calgary2012_PetrobrickVerificationGC.jpg|center|thumb|Figure 2: Gas Chromatograph demonstrating the differences in peak composition between an <i>E.coli</i> control and the Petrobrick. There was a large increase in a peak with a retention time of 12.25 min. suggesting that the Petrobrick was producing a new compound.|500px]]<html> | </html>[[File:Calgary2012_PetrobrickVerificationGC.jpg|center|thumb|Figure 2: Gas Chromatograph demonstrating the differences in peak composition between an <i>E.coli</i> control and the Petrobrick. There was a large increase in a peak with a retention time of 12.25 min. suggesting that the Petrobrick was producing a new compound.|500px]]<html> | ||
</html>[[File:Calgary2012_PetrobrickVerificationMS.jpg|center||thumb|Figure 3: Mass Spectra of the gas chromatograph peak at 12.25 min. The spectra suggests that the Petrobrick is selectively producing a C15 alkane. This is what was expected as determined by the Washington 2011 iGEM team.|500px]]<html> | </html>[[File:Calgary2012_PetrobrickVerificationMS.jpg|center||thumb|Figure 3: Mass Spectra of the gas chromatograph peak at 12.25 min. The spectra suggests that the Petrobrick is selectively producing a C15 alkane. This is what was expected as determined by the Washington 2011 iGEM team.|500px]]<html> | ||
| - | <p>With the Petrobrick shown to be able to successfully produce | + | <p>With the Petrobrick shown to be able to successfully produce |
| + | alkanes, it was time to test it out on naphthenic acids, to see if | ||
| + | |||
| + | they could be selectively converted into alkanes! This | ||
| + | |||
| + | experiment used commercially available naphthenic acid fractions | ||
| + | |||
| + | including a large number of different complex naphthenic acid | ||
| + | |||
| + | compounds. </p> | ||
<h2> Successful conversion of NA's into Hydrocarbons!</h2> | <h2> Successful conversion of NA's into Hydrocarbons!</h2> | ||
| Line 61: | Line 97: | ||
<h2><i>Nocardia</i> Carboxylic Acid Reductase (CAR)- Can we do better?</h2> | <h2><i>Nocardia</i> Carboxylic Acid Reductase (CAR)- Can we do better?</h2> | ||
| - | <p> | + | <p>Although we were successful using the Petrobrick to remove carboxyl groups from NAs, we wanted to improve on our results to see |
| + | |||
| + | if we could get a higher yield or possibly target other compounds. | ||
| + | |||
| + | One of our original fears in using the PetroBrick to | ||
| + | |||
| + | decarboyxlate naphthenic acids was that the first enzyme AAR was reported to be highly specific for fatty acids bound to ACP. We had concerns about its compatibility with naphthenic | ||
| + | |||
| + | acids and therefore sought another enzyme in the literature called | ||
| + | |||
| + | carboxylic acid reductase (CAR) that was documented to perform a | ||
| + | |||
| + | similar task as AAR, converting fatty acids to aldehydes, but with | ||
| + | |||
| + | much lower specificity (He et al, 2004). This enzyme, from <i>N. | ||
| + | |||
| + | iowensis</i> does not require covalent attachment to ACP so would | ||
| + | |||
| + | likely be much broader in substrate specificity. It requires a | ||
| + | |||
| + | second gene from <i>N. iowensis</i>, called Nocardia | ||
| + | |||
| + | phosphopantetheinyl transferase (NPT) necessary to append a 4’- | ||
| + | |||
| + | phosphopantetheine prosthetic group to CAR required for its full | ||
| + | |||
| + | function (Venkitasubramanian et al, 2006).</p> | ||
</html>[[File:Ucalgary Decarboxylation Team CAR Mechanism.jpg|center|350px|thumb|Figure 6. Mechanism of action of CAR]]<html> | </html>[[File:Ucalgary Decarboxylation Team CAR Mechanism.jpg|center|350px|thumb|Figure 6. Mechanism of action of CAR]]<html> | ||
| - | <p>Additionally, we had found | + | <p>Additionally, we had found another alternative: olefin-forming |
| + | |||
| + | fatty acid decarboxylase (OleT) from <i>Jeotgalicoccus sp. ATCC | ||
| + | |||
| + | 8456</i>. This is a decarboxylase of the cytochrome P450 family | ||
| + | |||
| + | that acts on fatty acids, but has also been documented to have low | ||
| + | |||
| + | substrate specificity (Rude et al, 2011). What was attractive | ||
| + | |||
| + | with this was that it was one single enzyme that go do the job of | ||
| + | |||
| + | the PetroBrick! Now that we knew that our decarboxylation approach | ||
| + | |||
| + | was valid, it was time to start testing and comparing this gene to | ||
| + | |||
| + | the PetroBrick</p> | ||
<h2> Where are we at? </h2> | <h2> Where are we at? </h2> | ||
| - | <p><i>N. iowensis</i> (NRRL 5646) | + | <p> We prepared genomic DNA from the host organism for CAR and NPT, <i>N. iowensis</i> (NRRL 5646) and successfully cloned the enzymes. CAR |
| + | |||
| + | was ligated into the PET vector and verified by a restriction | ||
| + | |||
| + | digest while NPT was cloned into psb1c3 (<a | ||
| + | |||
| + | href="http://partsregistry.org/wiki/index.php? | ||
| + | |||
| + | title=Part:BBa_K902061">BBa_K902061.</a>) and similarly | ||
| + | |||
| + | verified.</p> | ||
| + | |||
| + | <p>The reason for putting CAR into the pET47b+ plasmid was because of its six illegal cut sites for BioBrick | ||
| + | |||
| + | construction: one XbaI site, two EcoRI sites, and three NotI | ||
| + | |||
| + | sites. It needed to have these sites removed before it could be placed in an iGEM vector. These were first attempted using a multi-site | ||
| + | |||
| + | mutagenesis derived from the QuikChange® Multi Site Directed | ||
| + | |||
| + | Mutagenesis Kit, but this showed little success. Instead, a more | ||
| + | |||
| + | time-consuming but effective series of conventional single-site <a | ||
| + | |||
| + | href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/mutagen | ||
| + | |||
| + | esis">mutagenesis procedure</a> was favoured, using the KAPPA | ||
| + | |||
| + | amplification system. The XbaI and EcoRI sites were eliminated | ||
| + | |||
| + | first to that CAR could be moved from the pET Vector and ligated | ||
| + | |||
| + | into the PSB1C3 vector (<a | ||
| + | |||
| + | href="http://partsregistry.org/wiki/index.php? | ||
| + | |||
| + | title=Part:BBa_K902062">BBa_K902062.</a>). OleT was successfully | ||
| + | |||
| + | amplified from the <i>Jeotgalicoccus sp. ATCC 8456</i>. Like CAR, | ||
| + | |||
| + | OleT was inserted in a PET vector before placing it into a | ||
| + | |||
| + | BioBrick, as two illegal cut sites adjacent to one another needed | ||
| + | |||
| + | to be mutagenized. This part is now being ligated into psb1c3. | ||
| + | |||
| + | We are currently in the process of constructing all three parts | ||
| + | |||
| + | under contorl of a tetR promoter and ribosomal binding site (<a | ||
| + | |||
| + | href="http://partsregistry.org/Part:BBa_J13002">BBa_J13002</a>), | ||
| + | |||
| + | and then constructing these composite parts together as outlined | ||
| - | + | below.</p> | |
<h2>Final testing constructs</h2> | <h2>Final testing constructs</h2> | ||
Revision as of 13:31, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Decarboxylation

Why Decarboxylation?
Though there is great diversity within the naphthenic acids (NAs) class of compounds, all share the common chemical feature of a carboxylic acid group. The carboxyl group is the primary cause for their toxicity, allowing these chemicals to traverse cell membranes and react with cellular materials (Frank et al, 2009). NAs are recalcitranct (not easily degraded), potentially harmful to the surrounding ecosystem (Clemente & Fedorak, 2005) and corrosive to extraction and transport equipment of petroleum materials (Slavcheva et al, 1999). Corrosion of pipelines leads to higher maintenance costs as well as the grim possibility of these and other toxins leaking into the environment. There is a need for methods to degrade naphthenic acids that are not prohibitively expensive or that would result in production of other hazardous chemicals.
The main goal of OSCAR is to turn toxins like these into useable hydrocarbons by removing the carboxylic acid group(s) (Behar & Albrecht, 1984). Since naphthenic acids from petroleum deposits are a variable mixture, an enzymatic process with broad specificity is necessary. With the removal of the carboxylic acid moiety, we aim to produce alkanes suitable for use as fuel. The goal of this subproject was to find one or more suitable pathways to accomplish the decarboxylation of compounds such as naphthenic acids with the broadest specificity.
The PetroBrick
The 2011 Washington iGEM team developed the PetroBrick, a BioBrick consisting of two primary genes. These include acyl-ACP reductase (AAR), which reduces fatty acids bound to ACP to fatty aldehydes, and a second gene called aldehyde decarbonylase (ADC), which subsequently cleaves the entire aldehyde group and results in a hydrocarbon chain. Essentially this allows for hydrocarbons to be produced from glucose. What we realized though, is that the fatty acids that the PetroBrick targets, have a very similar structure to naphthenic acids.
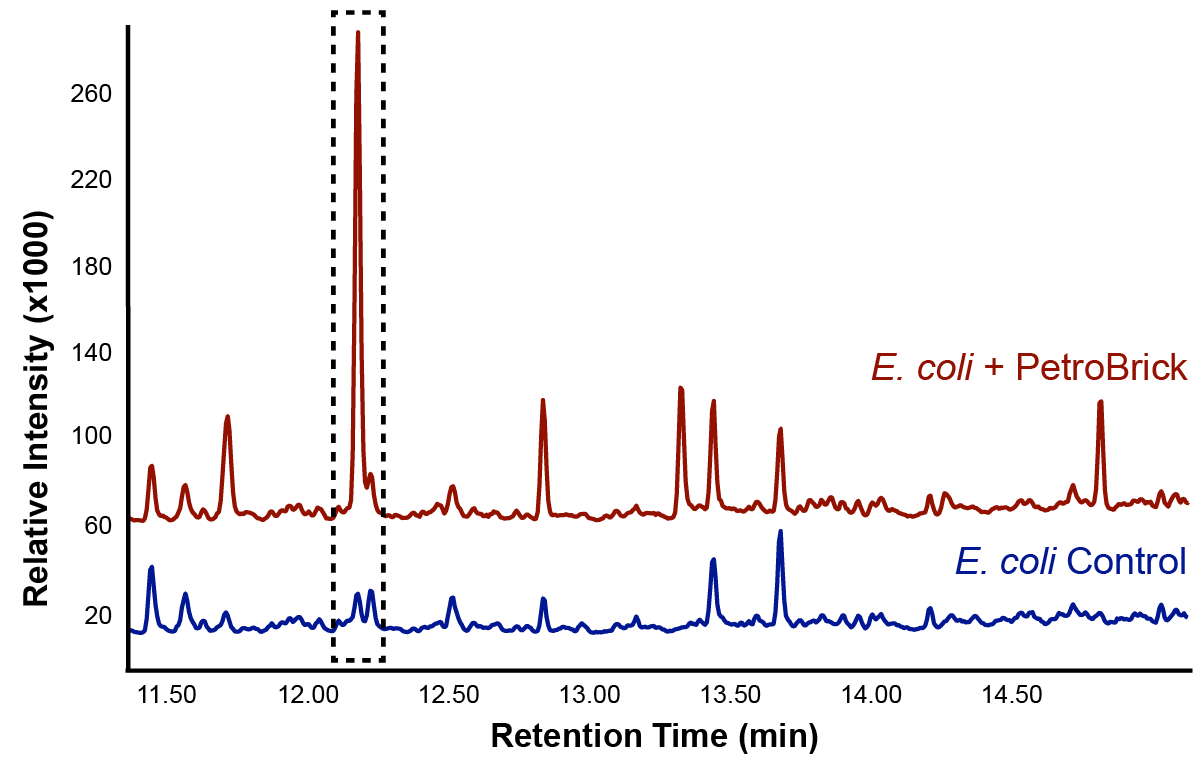
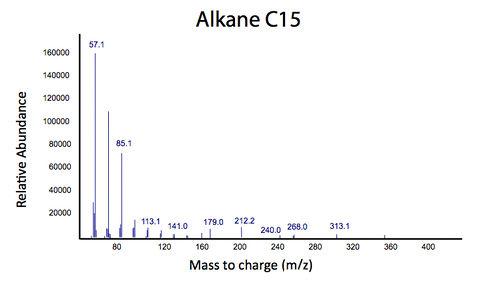
This lead us to believe that the PetroBrick may have the potential to turn naphthenic acids in to hydrocarbons and be a perfect solution to remediating naphthenic acids! First though, we needed to show that the PetroBrick did in fact work as expected. We had some difficulty with the DNA from the registry and had to request the constructs directly from the Washington team. Once we had the Petrobrick, we needed to verify that the Petrobrick would work in our hands as it did for the 2011 Washington team. Figures 2 and 3 demonstrate the function of the Petrobrick.
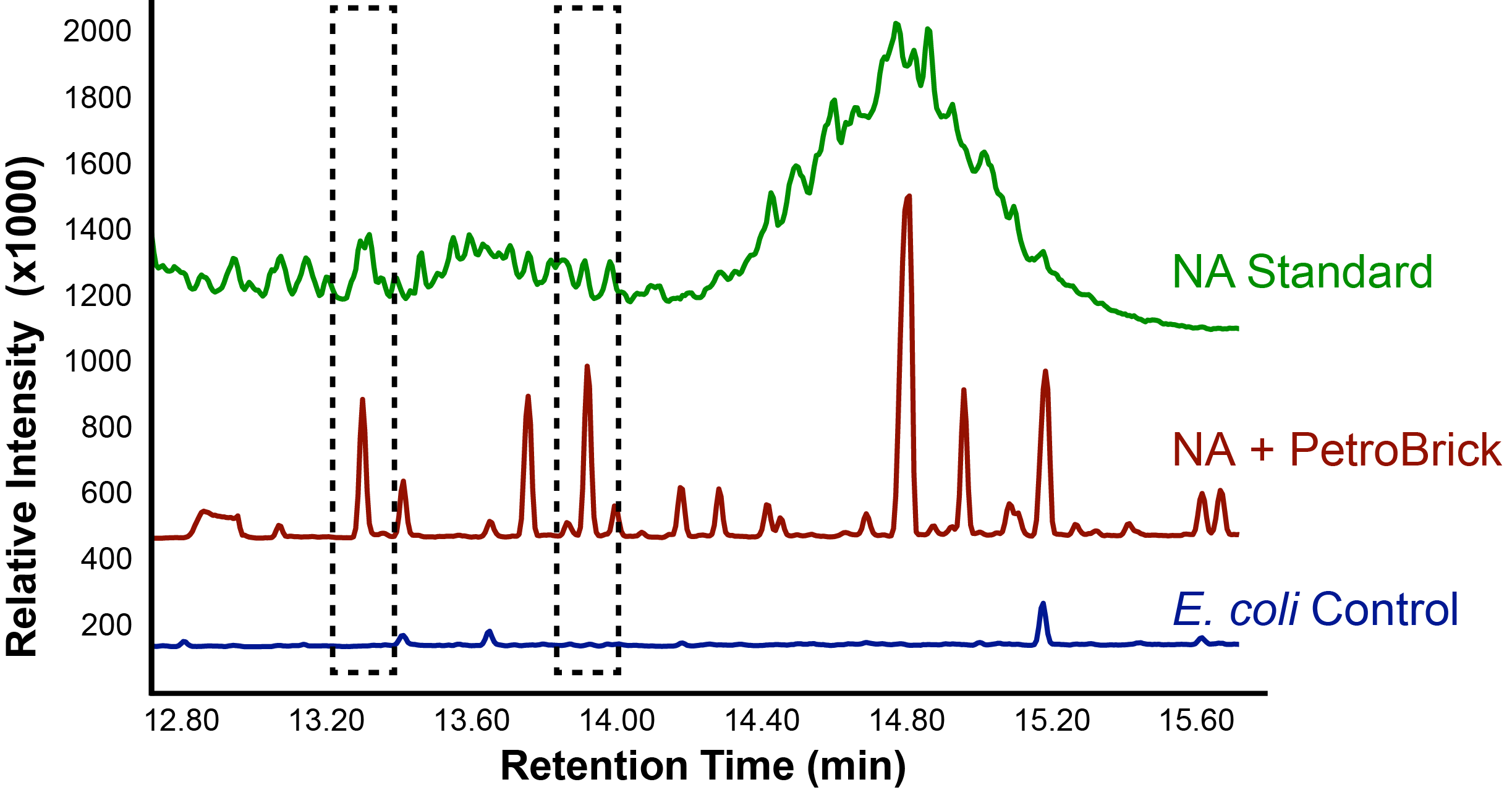
With the Petrobrick shown to be able to successfully produce alkanes, it was time to test it out on naphthenic acids, to see if they could be selectively converted into alkanes! This experiment used commercially available naphthenic acid fractions including a large number of different complex naphthenic acid compounds.
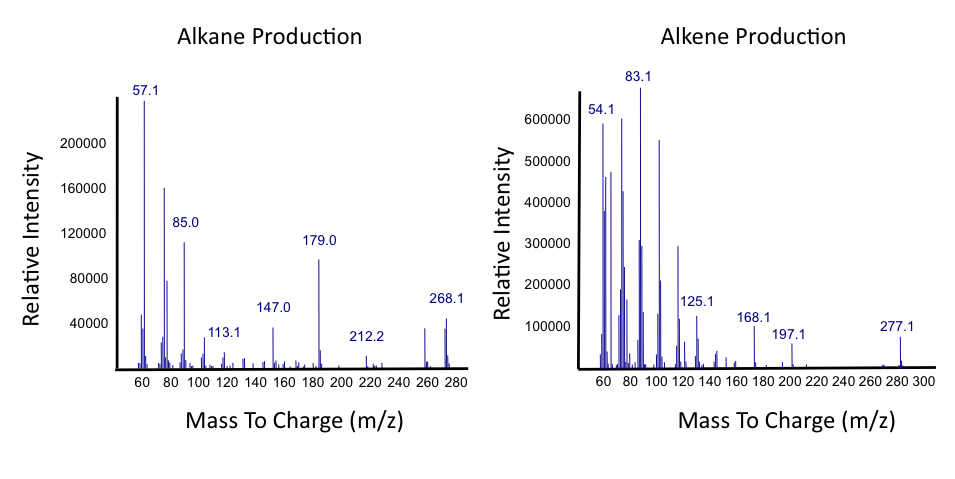
Successful conversion of NA's into Hydrocarbons!
The above graphs indicate that hydrocarbons were successfully produced from E.coli that contained the PetroBrick plasmid, as analysed with GC-MS. In Figure 2, E.coli containing the PetroBrick had significantly higher hydrocarbon peaks than in a control of E.coli that did not contain the PetroBrick plasmid. Not only was the PetroBrick able to degrade naphthenic acids into alkanes, but it was also able to produce alkenes as shown by Figure 3, indicating that the PetroBrick worked how we had expected it to!
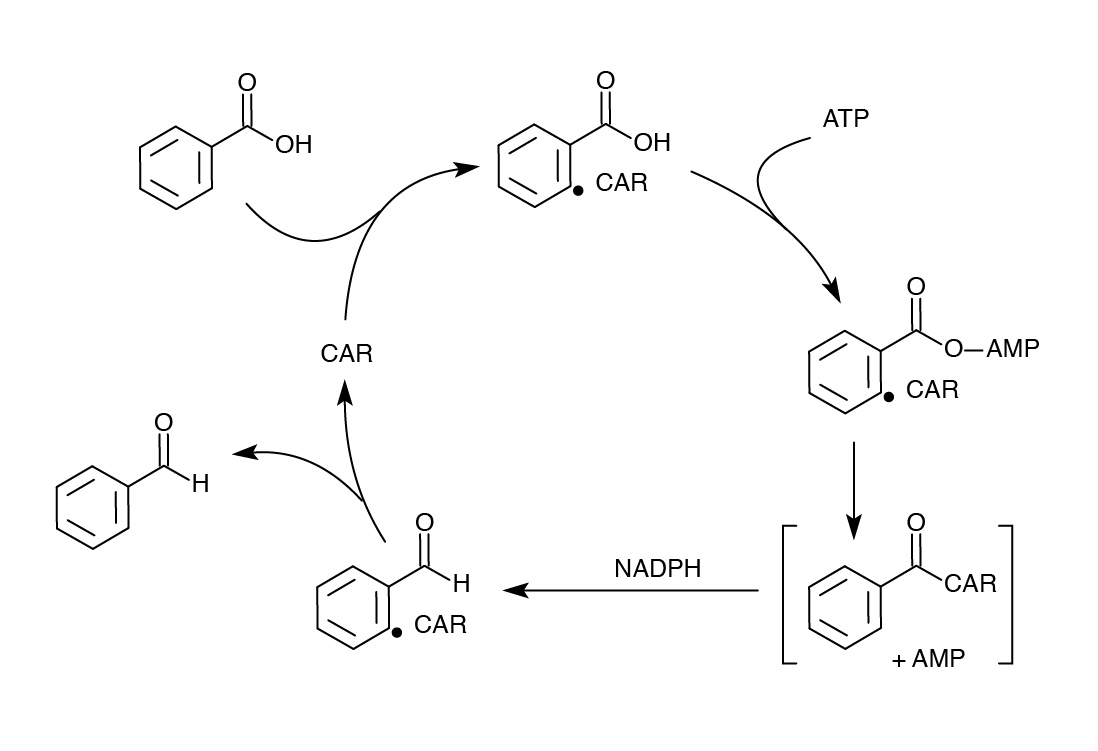
Nocardia Carboxylic Acid Reductase (CAR)- Can we do better?
Although we were successful using the Petrobrick to remove carboxyl groups from NAs, we wanted to improve on our results to see if we could get a higher yield or possibly target other compounds. One of our original fears in using the PetroBrick to decarboyxlate naphthenic acids was that the first enzyme AAR was reported to be highly specific for fatty acids bound to ACP. We had concerns about its compatibility with naphthenic acids and therefore sought another enzyme in the literature called carboxylic acid reductase (CAR) that was documented to perform a similar task as AAR, converting fatty acids to aldehydes, but with much lower specificity (He et al, 2004). This enzyme, from N. iowensis does not require covalent attachment to ACP so would likely be much broader in substrate specificity. It requires a second gene from N. iowensis, called Nocardia phosphopantetheinyl transferase (NPT) necessary to append a 4’- phosphopantetheine prosthetic group to CAR required for its full function (Venkitasubramanian et al, 2006).
Additionally, we had found another alternative: olefin-forming fatty acid decarboxylase (OleT) from Jeotgalicoccus sp. ATCC 8456. This is a decarboxylase of the cytochrome P450 family that acts on fatty acids, but has also been documented to have low substrate specificity (Rude et al, 2011). What was attractive with this was that it was one single enzyme that go do the job of the PetroBrick! Now that we knew that our decarboxylation approach was valid, it was time to start testing and comparing this gene to the PetroBrick
Where are we at?
We prepared genomic DNA from the host organism for CAR and NPT, N. iowensis (NRRL 5646) and successfully cloned the enzymes. CAR was ligated into the PET vector and verified by a restriction digest while NPT was cloned into psb1c3 (BBa_K902061.) and similarly verified.
The reason for putting CAR into the pET47b+ plasmid was because of its six illegal cut sites for BioBrick construction: one XbaI site, two EcoRI sites, and three NotI sites. It needed to have these sites removed before it could be placed in an iGEM vector. These were first attempted using a multi-site mutagenesis derived from the QuikChange® Multi Site Directed Mutagenesis Kit, but this showed little success. Instead, a more time-consuming but effective series of conventional single-site mutagenesis procedure was favoured, using the KAPPA amplification system. The XbaI and EcoRI sites were eliminated first to that CAR could be moved from the pET Vector and ligated into the PSB1C3 vector (BBa_K902062.). OleT was successfully amplified from the Jeotgalicoccus sp. ATCC 8456. Like CAR, OleT was inserted in a PET vector before placing it into a BioBrick, as two illegal cut sites adjacent to one another needed to be mutagenized. This part is now being ligated into psb1c3. We are currently in the process of constructing all three parts under contorl of a tetR promoter and ribosomal binding site (BBa_J13002), and then constructing these composite parts together as outlined below.
Final testing constructs
Final testing constructs are almost complete. These are illustrated in figure 7 and will allow us to compare the three different approaches. Unfortunately, as Washington only sent us the PetroBrick and not the two individual components, we will have to compare a combination of the PetroBrick and Car/NPT to the PetroBrick alone and to oleT.
 "
"