Team:Calgary/Notebook/Killswitch
From 2012.igem.org
Week 1 (May 1-4)
This was the first week where we met with other team members and summarized the primary subprojects the team will be tackling this coming summer.
Week 2 (May 7-11)
Members were assigned to the killswitch team. We spent a majority of this week performing literature searches and narrowing the killswitch to a few ideas.
The mechanism of death is still settled upon the micrococcal nuclease, but regulation of the death genes will be difficult. Existing repressible promoters in the registry still tend to be leaky in their expression, but we need to test this out. We may look into the TetR-repressible promoter (R0040) and also the clλ regulated promoter (R0051).
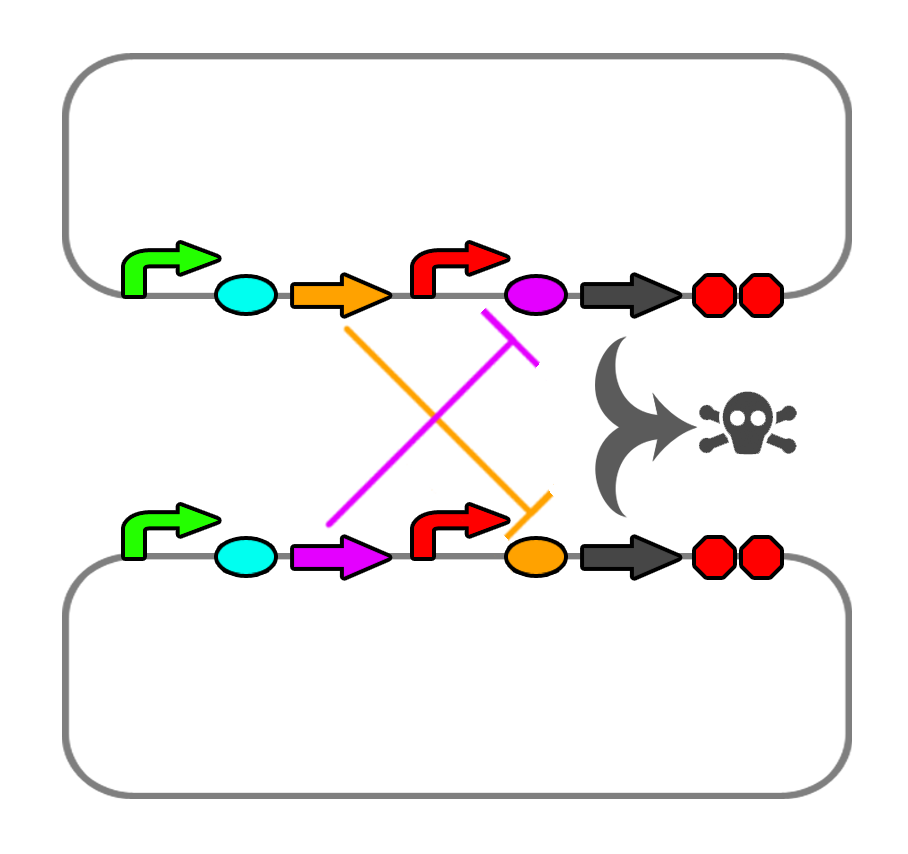
We also found a few riboswitches responsible to various metal ions such as Mg2+ and Mn2+. The mgtA riboswitch activates translation of the death gene when magnesium ions are not present in the solution. The mntA riboswitch deactivates translation of the death gene when manganese ions are not present. Considering this, we may be able to find a method of precipitating or otherwise sequestering Mn2+ ions out of the tailings water prior to entering the bioreactor. This way, our bacteria would not die when they come into contact with the manganese in the tailings water. Possible additives include carbonate (CO32-) or hydroxide (OH-), though this may alter the pH too greatly.
One other proposal would be to use promoters that activate when the bacteria detect the formation of a biofilm on, for example, glass beads. Engineering a monolayer of cells on a bead means that if a cell detaches, its death genes will activate. Further research into this is needed.
Week 3 (May 14-18)
We are continuing to look in to various other pathways. We discussed the possibility of a NOR-gated system to add a second layer of regulation to the kill gene. It would require the production of riboswitch ligands. Possible ligands include glucose, amino acids, and molybdenum cofactor.
We also looked into a co-dependent system where we would transform E. coli with two different plasmids and put them in the same bioreactor such that they produce ligands for each other that would repress the expression of the restriction enzyme and the nuclease. For this we explored riboswitches further and came up with the idea of using MOCO and SAM riboswitches as SAM is soluble in water and not found in the tailings ponds.
We also found a glucose repressible promoter that we could have potentially used upstream of the kill switch. This promoter is found in F. tularensis. However we also found that this promoter is not found in E. coli and F. tularensis has a unique polymerase which used this promoter. Hence using this promoter does not seem feasible. Next week we will be looking into further possibilities for the kill switch.
Week 4 (May 21-25)
In terms of our literature search we decided against the glucose repressible promoter from F. tularensis because it was not native to E.coli and upon contact with the authors of the paper we found that the promoter did not work in E.coli. We also came upon two systems that we are interested in further investigating. The first one was a glucose activated promoter that was found in the registry which we thought of coupling to an inverter and the second one is a rhamnose-based regulatory system.
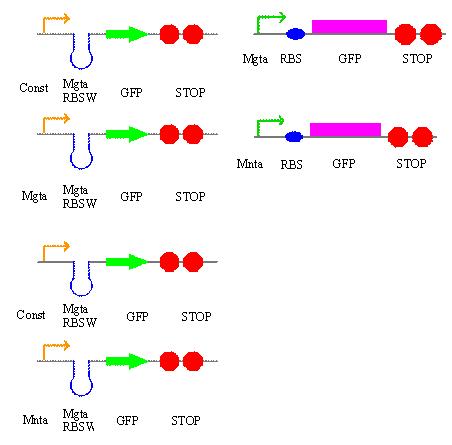
On Tuesday, in addition to our literature research, we decided on some of the circuits that we will be building over the next couple of weeks and came up with a plan detailing what will be done each day. These are the circuits:
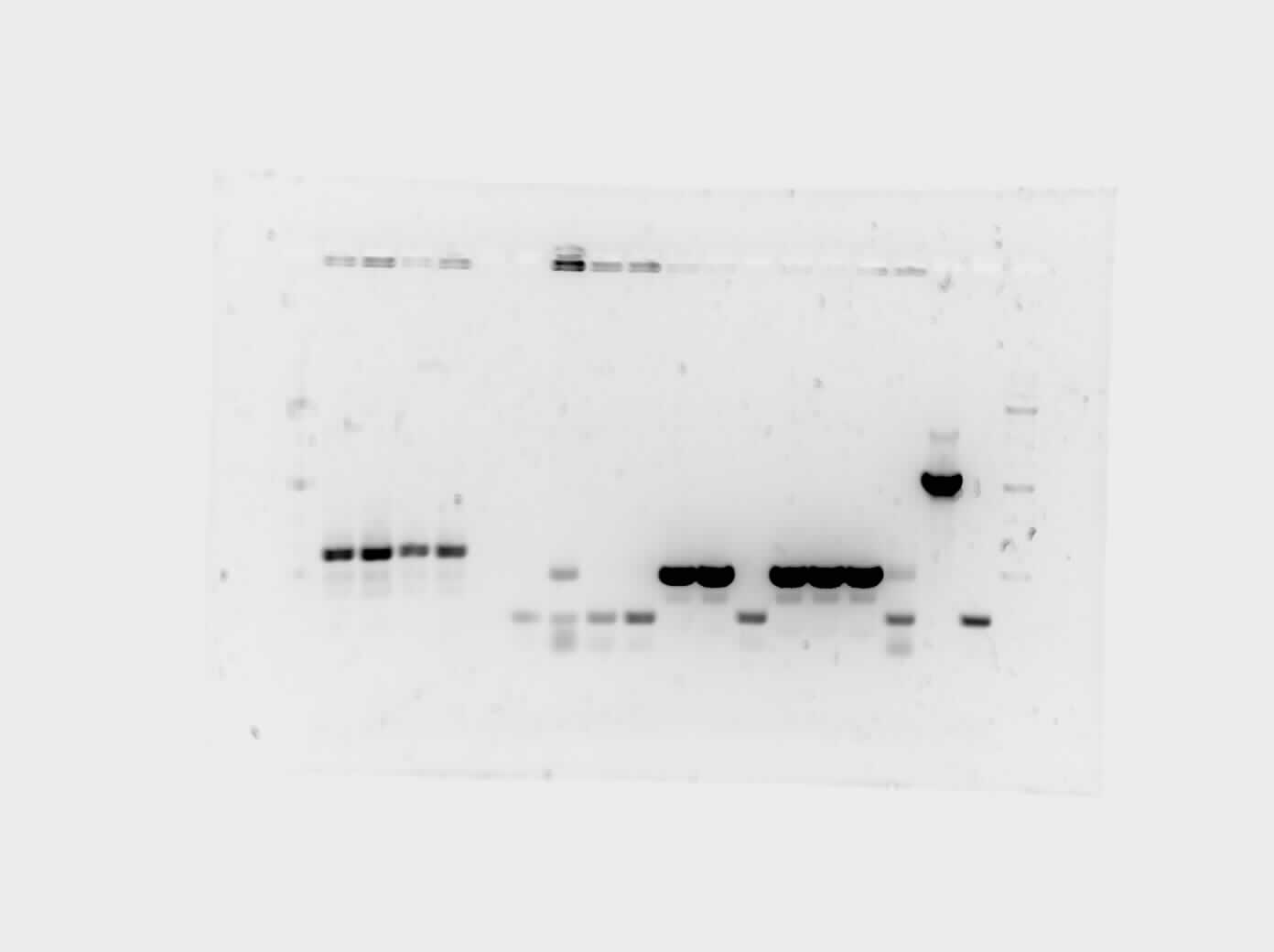
On Wednesday we proceeded with the transformation of 3 parts from the iGEM kit into our E.coli: a tetR promoter, a GFP-double terminator, and a RBS-GFP-double terminator. On Thursday we verified our transformation process with a cPCR where we ran 10 colonies from each plate with our controls. We ran a gel on the PCR products and viewed it to verify the transformation and to decide which of the successful colonies to plate and grow overnight.
On Friday we isolated the plasmids through a mini prep. The plan for next week is to do a restriction digest and run it on a gel to confirm that the plasmids have the part. We also want to start more transformation of our parts into E.coli such as the riboswitches and the promoters. We want to verify the transformation, isolate the plasmids and verify, and start the DNA construction digest to start building our circuit.
 "
"