Team:Calgary/Notebook/CatecholDegradation
From 2012.igem.org
| Line 60: | Line 60: | ||
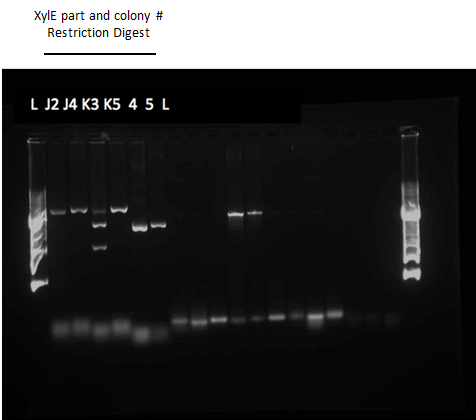
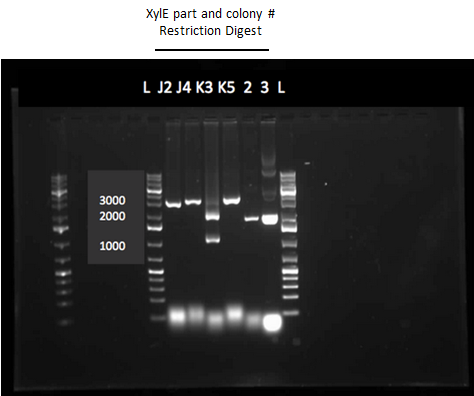
</html>[[File:UCalgary2012Week12agelXylE.PNG|500px|thumb|XylE Restriction Digest Gel. Both XylE parts from the registry were transformed into E.coli. One part contained the XylE gene and a native RBS site (J33204) and the other contained glucose repressible promoter (Pcst) and the XylE gene and a native RBS site (K118021). After running a colony PCR on the transformation plates two colonies from each plate looked like they contained the XylE gene. Overnight cultures of these colonies were made and restriction digests were ran on the mini-prepped cells. Lanes 2 and 3 contain the restriction digest product from colonies 2 and 4 on the J33204 plate and lanes 4 and 5 contain the product from colonies 3 and 5 from the k118021 plate. The two bands in 3rd lane after the 1 Kb plus ladder indicate that the restriction digest for that plasmid was successful. The top band around 2000 base pairs represented the pSB1A3 vector (2155 bp) and the bottom band around 1000 base pairs represented the XylE gene (1097 bp).|center]] | </html>[[File:UCalgary2012Week12agelXylE.PNG|500px|thumb|XylE Restriction Digest Gel. Both XylE parts from the registry were transformed into E.coli. One part contained the XylE gene and a native RBS site (J33204) and the other contained glucose repressible promoter (Pcst) and the XylE gene and a native RBS site (K118021). After running a colony PCR on the transformation plates two colonies from each plate looked like they contained the XylE gene. Overnight cultures of these colonies were made and restriction digests were ran on the mini-prepped cells. Lanes 2 and 3 contain the restriction digest product from colonies 2 and 4 on the J33204 plate and lanes 4 and 5 contain the product from colonies 3 and 5 from the k118021 plate. The two bands in 3rd lane after the 1 Kb plus ladder indicate that the restriction digest for that plasmid was successful. The top band around 2000 base pairs represented the pSB1A3 vector (2155 bp) and the bottom band around 1000 base pairs represented the XylE gene (1097 bp).|center]] | ||
[[File:UCalgary2012Week12bgelXylE.PNG|500px|thumb|Restriction digest gel of XylE gene with rbs site (J33204). Lanes 10-14 (Colonies A-J). Successfully digested plasmids from colonies B and D are at an expected size of 1000 bp with a 1Kb ladder (L).|center]] | [[File:UCalgary2012Week12bgelXylE.PNG|500px|thumb|Restriction digest gel of XylE gene with rbs site (J33204). Lanes 10-14 (Colonies A-J). Successfully digested plasmids from colonies B and D are at an expected size of 1000 bp with a 1Kb ladder (L).|center]] | ||
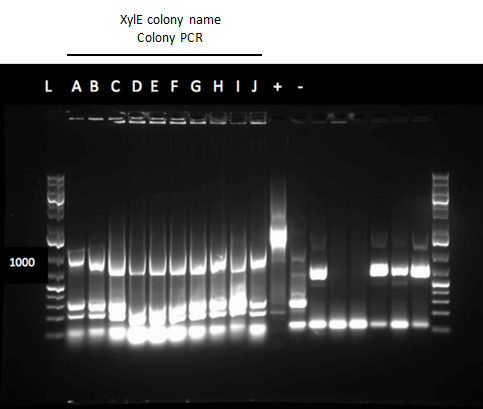
| - | [[File: | + | [[File:UCalgary2012Week12cgelXylE.PNG|500px|thumb|Colony PCR gel of XylE with rbs site (J33204). Lanes 9-13 (Colonies A,B,D,J) with a 1Kb ladder (L).|center]]<html> |
Revision as of 05:38, 2 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Decatecholization Journal
Week 1 and 2 (May 1-4 and May 7-11)
The Catechol degradation part of the project started off as general ring cleavage. The goal was to cleave both the aromatic and aliphatic rings contained in many toxic compounds in the tailings ponds. The first two weeks involved getting familiar with lab procedures and researching aromatic ring cleaving using intra- and extradiol dioxygenases from species of Pseudomonas and Bacillus. We also began literature searches on aliphatic ring cleavage done by monooxygenases. The project was focused on ring cleavage from week 1-12 and catechol degradation was focussed on in the following weeks.
Week 3 (May 14 - May 18)
In the second week we continued to do more research on the degradation of alicyclic compounds and found two strains of bacteria that contained genes needed for this process. The first strain, Thauera butanivorans, contains the genes required to activate the ring by adding a hydroxyl group. The gene is called butane monooxygenase and is composed of three subunits, a hydroxylase, a reductase, and a regulatory component. The second strain, Acinetobacter sp. SE19, contains the genes needed to oxidize the alcohol, formed by butane monooxygenase, and to cleave the ring. There is a cluster of nine genes that perform this oxidation and cleavage but only six are involved directly.
Week 4 (May 22 - May 25)
We started looking into more organisms that can carry out cyclohexane degradation and found one called Brachymonas petroleovorans. This organism is capable of both hydroxylating cyclohexane and carrying out cyclohexanol oxidation. We are also hoping to find evidence that Pseudomonas fluorescens, Pf-5 has genes that can cleave aliphatic rings, however, first we have to determine if Pf-5 can grow in the presence of butylcyclohexane. To do so we carried out an assay, using LB, testing the toxicity of butylcyclohexane on Pf-5. After 24 hours of incubation we took OD600 readings¬ which suggested that the presence of butylcyclohexane did not have effect on cell growth. We also looked up Pseudomonas fluorescens Pf-5 in the Pseudomonas Genome Database and found genes that code for proteins with similar functions to what we have found for aromatic and aliphatic ring cleavage.
Week 5 (May 28 - June 1)
This week we redid the previous assay with Pf-5 and butylcyclohexane using glass vials instead of plastic falcon tubes. These vials allowed for more surface and better containment of the volatile compound. We carried out this assay using both LB and M9-MM (M9 minimal media). After 24 hours we took OD600 readings and obtained similar results as last week. The growth was considerably decreased in the M9-MM, however we are not sure whether the bacteria were able to use the compound as a carbon source because the MM contained glucose. The fact that the growth decreased showed that they might depend on the glucose in the media to grow.
Week 6 (June 4 - June 8)
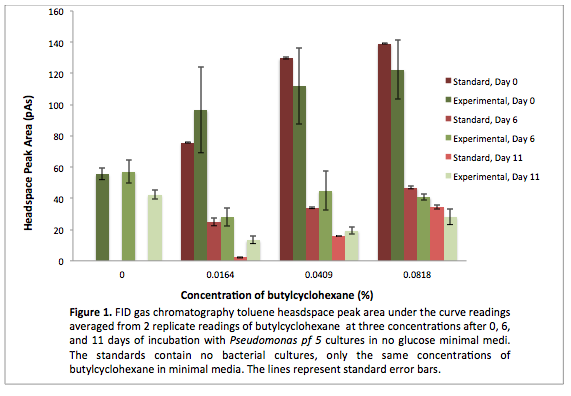
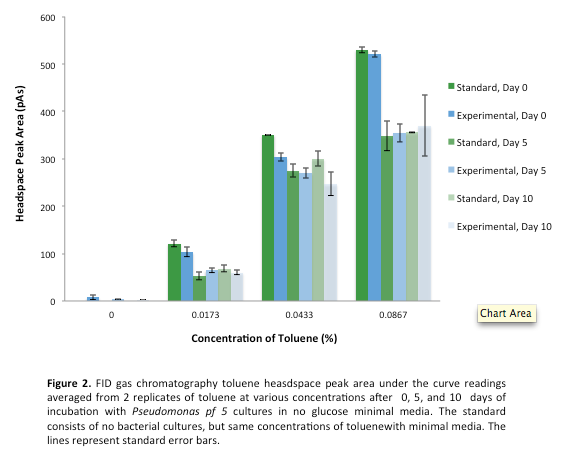
This week we made subcultures from Pf-5 in M9-MM into M9-MM without glucose to avoid carry-over of glucose, to insure that the bacteria use butylcyclohexane as their only carbon source. We also took GC (Gas Chromatography Mass Spectroscopy) readings and compared experimental values to standards made with water and butylcyclohexane (butCH). Readings showed a decrease in amount of butCH in the headspace, compared to standards, indicating that the compound was being degraded by Pf-5. From these cultures we subcultured Pf-5 in M9-MM without glucose into the same media and butCH, and marked this reading as our zero time point. Furthermore, we made subcultures from original Pf-5 in M9-MM without glucose into M9-MM and toluene to test for pathways that degrade aromatic structures. To identify possible genes for the pathways we started running blasts on the Pf-5 genome looking for genes that are homologous to ones that we have been looking at for the two ring cleavage pathways (aromatic and aliphatic). In addition we started a verification of XylE (aromatic ring cleavage enzyme) by transforming part K118021 (XylE with a Pcst promoter) from the parts registry and did two colony PCRs. Unfortunately none of them showed successful results and only the positive control showed an appropriate length DNA.
Week 7 (June 11 - June 15)
Since last week we continued taking GC readings on the Pf-5 incubated with butylcyclohexane and toluene. At this point the Pf-5 had been incubated with butylcyclohexane for 6 days and with toluene for 5 days. The amount of both butylcyclohexane and toluene decreased by a large amount in the experimental cultures but the amount of each in the abiotic standards also largely decreased by about the same amount. This lead us to conclude that the reason for decrease was not due to the bacteria metabolizing the compound, but due to unknown abiotic factors. To measure relative amounts of bacterial growth we took OD600 readings for both cultures which were very low and indicated that there was little bacterial growth. The verification of XylE from the parts registry was continued using a miniprep on three overnight cultures from part K118021. The DNA concentrations were very high likely due to genomic contamination of the miniprep. A second part (J33204) containing XylE and an rbs site was transformed and a colony PCR was done on part J33204 but the gel did not indicate successful results, and again only the positive control worked. No positive colonies were found.
Week 8 (June 18 - June 22)
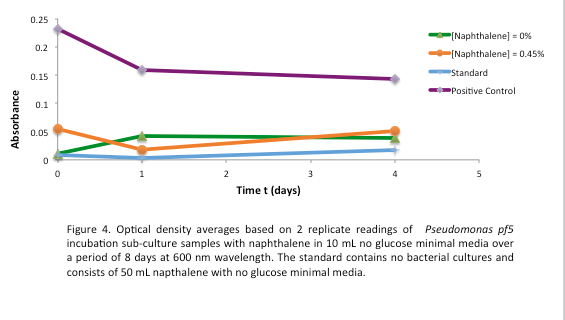
This week we started a new assay where Pf-5 was incubated with naphthalene. Since naphthalene is not volatile, GC headspace reading was not an appropriate method of measurement to indicate decrease of the compound. Instead we took OD600 readings to measure bacterial growth. The original cultures were taken from a Pf-5 culture in LB and inoculated into M9-MM without glucose. After 24 hours of growth these were subcultured into fresh M9-MM without glucose, again to prevent carryover of glucose from the LB. Positive controls were also made that consisted of M9-MM with glucose and Pf-5. OD600 readings of the subcultures were taken over a period of 8 days. GC readings were done on the Pf-5 incubated with butylcyclohexane after 11 days and with toluene after 10 days. The trend remained the same as the measurements done after 5 and 6 days in the previous week. The amount of compound decreased in the experimental cultures but the amount of compound also decreased in the abiotic standards almost in equal proportions. With the results after 10 and 11 days we could see that the decrease was not due to the bacteria and we decided that the strain we were using did not contain the genes necessary for butylcyclohexane or toluene degradation. Throughout the week we continued the verification of XylE by performing a second colony PCR from the transformation plate using part J33204. The gel did not contain DNA of the right lengths and only the positive control worked.
Week 9 (June 25 - June 29)
After the 8 day incubation the readings of the naphthalene assay were graphed and there was a definite increase in the cell growth of the cultures containing naphthalene as a carbon source which indicated that the bacteria were most likely metabolizing the compound. A verification of another part, Alkane hydroxylase (alkB2rubBrubA3rubA4) system, from the registry was started. The system consists of 4 genes that have been shown to hydroxylate C5-C8 cycloalkanes. We chose this part to assist in the degradation of cyclohexane and cyclopentane. The transformation was successful and the gel of the colony PCR showed one positive colony but since there was contamination in the gel we were not sure if we could conclude that the PCR had worked. Final GC-MS readings on the butCH and toluene cultures were taken and results were graphed.
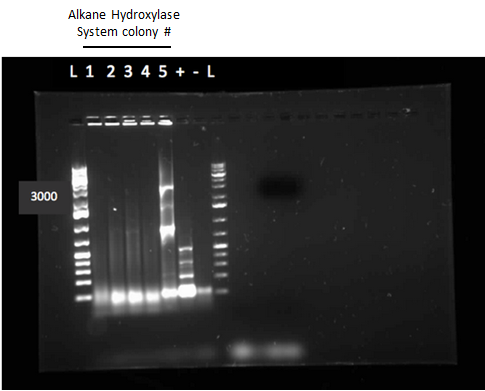
Week 10 (July 2-July 6)
This week we performed another colony PCR of 5 colonies from the transformation plate of part K398014 (alkane hydrolase construct: alkB2rubBrubA3rubA4) and ran the products on a gel. We obtained 4 positive colonies and made overnight cultures. We decided to stop using this part for the project and therefore did not require a further miniprep on these overnight cultures. However we made an overnight culture of the positive colony from the alkane hydroxylase system gel (colony 5) and miniprepped the overnight culture. This resulted in a good concentration of 345.6 ng/μL as we verification of XylE by doing two more colony PCRs of each part (K118021 and J33204) but only the positive control worked when we ran the gel.
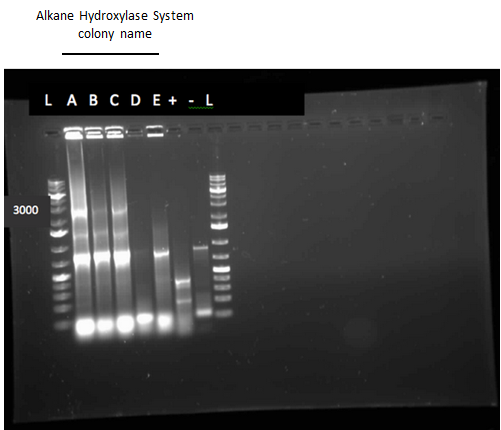
Week 11 (July 9 - July 13)
Since we could not obtain any positive colonies from last weeks colony PCRs we began the transformation of each part again. The transformations were successful and when the colony PCR products were run on a gel there were two positive colonies for each part. Overnight cultures of the positive colonies were made. Minipreps were done on the overnight cultures and a restriction digest was performed on the products of the minipreps. The restriction digest products were run on a gel and there was one that looked like it had been successful but due to a poor quality ladder on this gel we could not make any final conclusions on the success of the restriction digest.
Week 12 (July 16 - July 20)
We began the week by rerunning the restriction digest products on a gel with a good ladder and obtained one positive result with two bands. One band was around 1000 bp for the part and the other was at about 2000 bp for the plasmid backbone. We made a streak plate of the colony that had worked for the restriction digest and sent the miniprep product for sequencing. The colony that worked was from the transformation plate of part K118021. We also repeated a colony PCR of the transformants from part J33204 using 10 colonies and all of them looked like positive colonies. Overnight culutres were made and a miniprep and restriction digest was completed.
Week 13 (July 23 - July 27)
The colony PCR of part J33204 (XylE with a native rbs site) was done again and all five colonies looked like they contained the plasmid after running the products on a gel. After miniprepping and running a restriction digest on these five colonies two of them were sent to sequencing. An assay using the compound catechol was also started this week. E.coli cells (top 10) transformed with the part K118021 (XylE with a Pcst promoter) were used in this assay. Six overnight cultures were made, 3 using M9-MM and 3 using LB. These were spun down and resuspended in fresh M9-MM and LB and brought to a concentration of 0.1 M catechol by using a 1M stock solution. There seemed to be an initial colour change in the reactions from clear to yellow which was only visible in the cultures using M9-MM. All of the cultures were incubated overnight and they turned a black green colour. For this assay we should have used the supernatant because the reaction takes place outside of the cell so this is what we did next week.
Week 14 (July 30 - August 3)
The sequencing results for the part J33204 matched the part so a construction was started. The construction was a 3-way ligation putting the tetR promoter (R0040) and XylE (J33204) into a pSB1C3 vector. The ligation product was transformed into competent cells and a colony PCR was done on the transformants using both bio brick and R0040 primers. The primers for XylT, which is a ferredoxin that allows for XylE to be activated after a reaction with catechol, arrived and a colony PCR on four different strains of Pseudomonas putida was done. None of the strains seemed to contain the TOL plasmid, which is where XylT is found in certain strains of P. putida, because no bands showed up in the gel. The catechol assay was continued this week. Six overnight cultures were made, 3 using M9-MM and 3 using LB. They were spun down and the supernatant was brought to a catechol concentration of 0.1M, 0.2 M and 0.5 M using a 1M stock solution. The solutions turned light yellow initially, but after a few hours they turned brown. The assay was repeated with the same procedure as above but a control, a colony that did not contain the part K118021, was added. After the catechol was added to the supernatants there was a colour change from clear to yellow in all of the tubes, even the control. For a negative control catechol was added to M9-MM and this solution became pink. The tubes were incubated on the bench and checked regularly. The colour changed from yellow to pink to brown when they were left on the bench overnight.
Week 17 (August 20 - August 24)
The Catechol assay was repeated with a new procedure. Overnight cultures were made in LB and washed the pellet in M9-MM without glucose for varying amounts of time. These cultures were spun down and the supernatant was brought to a concentration of 0.1 M. All of the supernatants became bright yellow except for the control which did not contain the K118021. The colour change indicated that catechol was being converted to 2-HMS (2-Hydroxymuconate semialdehyde). The three way ligation didn’t work so a two-way ligation with R0040 (tetR) in a pSB1A3 vector and J33204 (XylE) in a pSB1C3 vector. One reaction was done where R0040 was the vector and J33204 was the insert, and another reaction was done where R0040 was the insert and J33204 was the vector. Both ways worked and the new procedure for the catechol assay was done with the transformed cells and all of the supernatants turned bright yellow.
Week 18 (August 27 - August 31)
The sequencing results of the ligation were positive so assays were started with the cells transformed with the construct R0040-XylE. The assays involved the use of the petrobrick and a species of micrococcus. The purpose of these experiments was to see if any of the oxygen groups on the product of catechol degradation, which is 2-HMS, were removed.
 "
"