Team:CD-SCU-CHINA/Notebook
From 2012.igem.org
| Line 1: | Line 1: | ||
{{SCU-IGEM}} | {{SCU-IGEM}} | ||
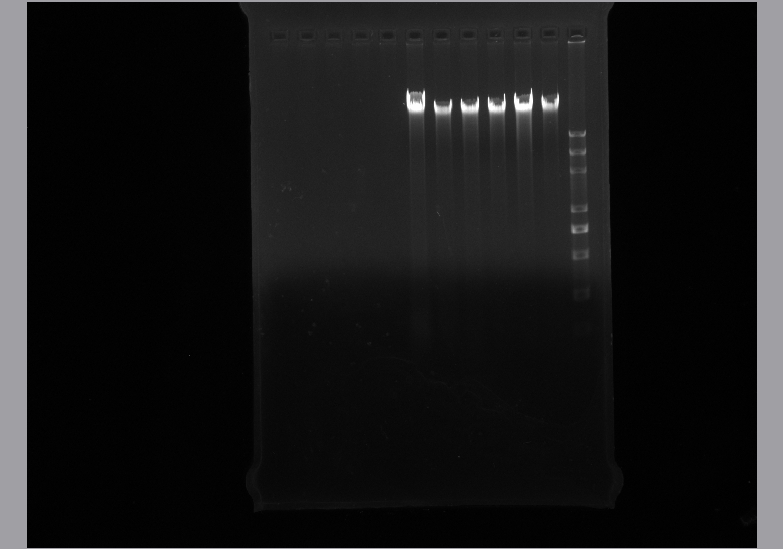
* '''Genome extraction''' | * '''Genome extraction''' | ||
| - | ** Using gene extraction kit to isolate the whole genome of Alcanivorax borkumensis SK2 | + | ** Using gene extraction kit to isolate the whole genome of Alcanivorax borkumensis SK2<br> |
| + | [[File:Genome 8.5.jpg]] | ||
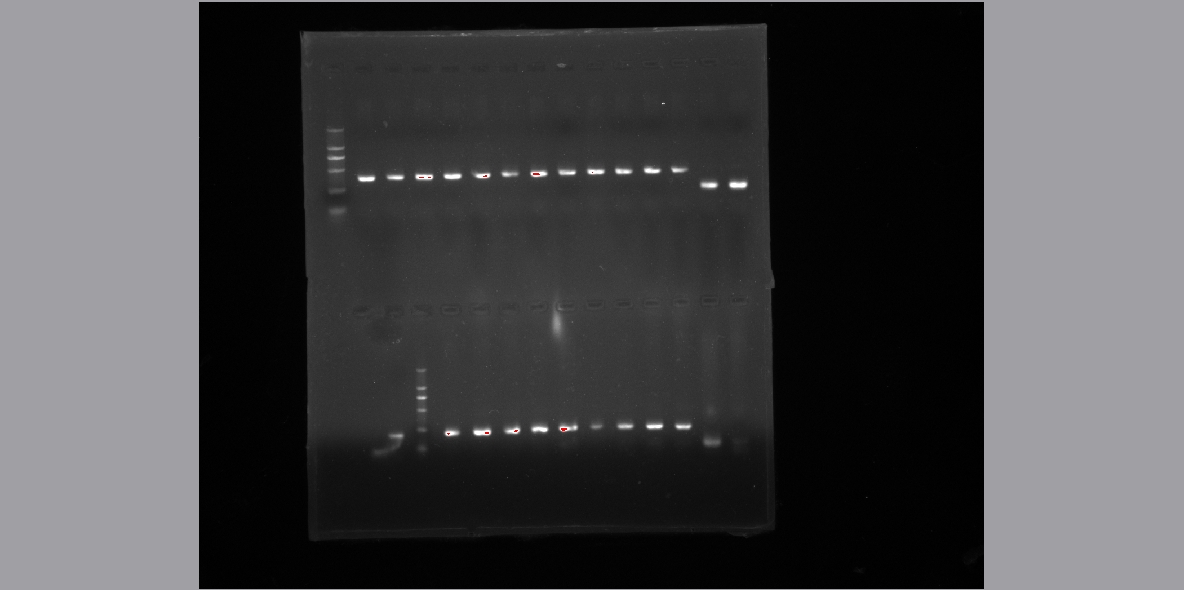
* '''Gene isolate using PCR''' | * '''Gene isolate using PCR''' | ||
| - | ** Normal primer design and '''gradient PCR''' to amplify the gene. | + | ** Normal primer design and '''gradient PCR''' to amplify the gene.<br> |
| + | [[File:Gradent2.jpg]] | ||
* '''Site mutation''' | * '''Site mutation''' | ||
** Using the primer design by us, follow these tips: | ** Using the primer design by us, follow these tips: | ||
| Line 17: | Line 19: | ||
Gne inserts: more than 5:1 molar ratio to vector | Gne inserts: more than 5:1 molar ratio to vector | ||
In-fusion enzyme: 2ul every 10ul reaction system | In-fusion enzyme: 2ul every 10ul reaction system | ||
| - | ddH2O: Add water to 10ul | + | ddH2O: Add water to 10ul<br> |
| - | + | [[File:Colony.jpg]] | |
* '''Sequensing''' | * '''Sequensing''' | ||
Done by company | Done by company | ||
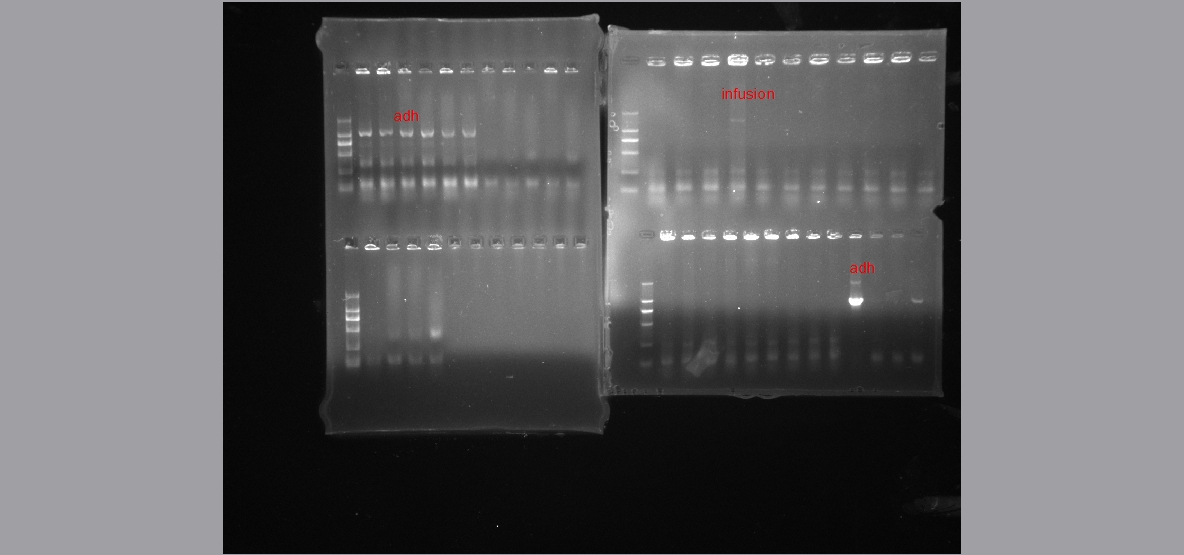
* '''Colony PCR Plasmid extraction''' | * '''Colony PCR Plasmid extraction''' | ||
| - | Colony PCR sometimes was not as convenient as we thougt. | + | Colony PCR sometimes was not as convenient as we thougt. Sometimes plasmid extraction is better! |
| + | [[File:Plasmid ex.jpg]] | ||
* '''Double Digestion for chek''' | * '''Double Digestion for chek''' | ||
* '''Temperament chromatography''' | * '''Temperament chromatography''' | ||
for the function test of the degradation of the two enzyme gene. | for the function test of the degradation of the two enzyme gene. | ||
Latest revision as of 03:42, 27 September 2012
- Genome extraction
- Using gene extraction kit to isolate the whole genome of Alcanivorax borkumensis SK2
- Using gene extraction kit to isolate the whole genome of Alcanivorax borkumensis SK2
- Gene isolate using PCR
- Normal primer design and gradient PCR to amplify the gene.
- Normal primer design and gradient PCR to amplify the gene.
- Site mutation
- Using the primer design by us, follow these tips:
1,Full length not less than 28bp; 2, 3'end to mutation site not less than 18bp; 3, Mutation site to 5' end not less than 10bp.
- Gene purification ,Enzyme digest, ligation and transformation
In our experiment, we found the vector (we used) is hard to digest by two enzyme in the same time, it cost us a long time on the identification the correct plamid, unfortunately, it was frustrating. at last, we follow the advice from the teacher, degest one by one, and have an self-ligation test in order to have the correct digested vector.
- In-fusion cloning and tranformation
we use this method for the ligation of 6 genes, but follow the instrction of the Clontech company, we may waste a lot of money on it, so we search methods from the paper, first, we test four-way ligation, however, it turn out to be no colony. so we test three-ligation. it was useful.
Methods are as follows:
Vector: more than 50ng;
Gne inserts: more than 5:1 molar ratio to vector
In-fusion enzyme: 2ul every 10ul reaction system
ddH2O: Add water to 10ul
- Sequensing
Done by company
- Colony PCR Plasmid extraction
Colony PCR sometimes was not as convenient as we thougt. Sometimes plasmid extraction is better!
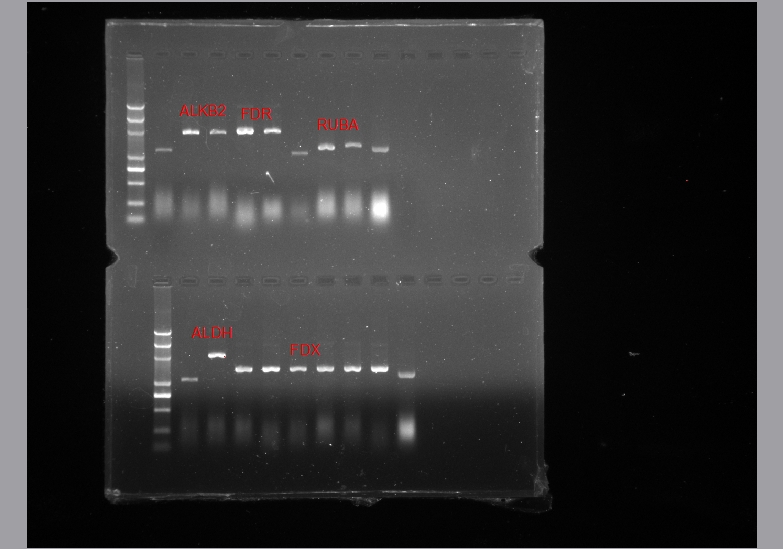
- Double Digestion for chek
- Temperament chromatography
for the function test of the degradation of the two enzyme gene.
 "
"