Team:Buenos Aires/Results/Bb2
From 2012.igem.org
(→Terminator) |
(→Terminator) |
||
| Line 116: | Line 116: | ||
{| class="wikitable" border="1" | {| class="wikitable" border="1" | ||
|- | |- | ||
| - | |<!--column1-->[[File:Bsas2012-Translate.png| | + | |<!--column1-->[[File:Bsas2012-Translate.png|500px]] |
|- | |- | ||
|Picture: General Design of Biobrick 5. | |Picture: General Design of Biobrick 5. | ||
|} | |} | ||
Revision as of 16:53, 24 September 2012

Contents |
Construct Desing based on Biobrick Registry Parts
His Secretion in Bacteria In the spirit of the competition we decided to design an extra construct besides our main biobrick. In this case, we decided to create it solely by using parts that came in the iGEM Kit 2012 distribution, and that could work for the same purposes that our main biobrick, which is mainly the export of aminoacids.
We designed a plausible construct that could work for the export of His using 5 parts of the registry, but we didnt find enough parts in order to design a simmilar one for the export of Trp.
Furthermor, the binding and preparation of this device is much more complex and has many more steps than what our main biobrick needs to work.
Therefore, we conclude that our main biobrick is an important contribution to the registry part, given that it allows the export of aminoacids to be enhanced with the use of only one part, without the need of the many steps that we describe in this section and consequently reducing the risk of failure and errors.
Aim
To create a biobrick that would enhance Histidine secretion in E. coli using standard parts of the registry as a proof that one can increase the production and secretion of an aminoacid and its measurement in the culture medium.
To learn how to use and merge standard parts from the registry provided at the iGEM Kit.
To characterize the functioning of existent parts in the registry and new combinations of them, therefore contributing the improvement of the iGEM record.
To proove that our main biobricks, devices 1 and 3 for the export of His, are an important contribution to iGEM, given that they are capable of His export with far less steps involved.
To proove that our main biobricks, devices 2 and 4 for the export of Trp, are an important contribution to iGEM, given that there are was no biobrick with such function available.
Parts to use
In order to accomplsPromoter (constitutive or inducible); RBS; Peptide Signal (Secretion Tag), Histidine Tag (repeated) and a Terminator.
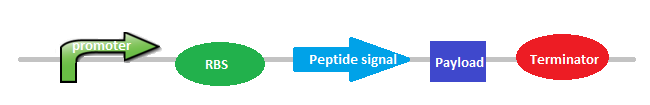
|
| Picture: General Design of Biobrick 5. |
Promoter choice
We found many usable parts to use as promoters. We found Promoters + RBS ideal for our purposes, in order to economize ligation steps.
We considered using two Biobricks:
1) Promoter + RBS: BBa_K206015 Strongest constitutive promoter in J23100 family (J23100) + mid-strength RBS from the community collection (B0030, 0.6) It looks as a reliable sequence but it has not been tested according to the resgistry.
2) Inducible Promoter (IPTG) + RBS (Strong): BBa_J04500
This part has been tested and according to the registry it works well.
We finally decided to use Biobrick 2: BBa_J04500, in order to make our system plausible of regulation through IPTG.
This kind of regulation could also have been implemented in our main biobricks 1 and 3, for the same purposes or making the system more flexible.
Signal Peptide Options
Unfortunately, we found very few signal peptide biobrick options, solely two and tested in Cyanobacterium.
Our two options were:
1) pilA1 signal sequence from cyanobacterium Synechocystis; secretes protein: BBa_K125300
Sequence:
tggctagtaattttaaattcaaactcctctctcaactctccaaaaaacgggcagaaggtggt
2) slr2016 signal sequence from cyanobacterium Synechocystis; secretes protein: BBa_K125310
Sequence:
tggcagcaaaacaactatggaaaattttcaatcctagaccgatgaagggtgga
These parts are only partically confirmed and optimized for working in Cyanobacterium, not E. coli or Yeast. We could use any of them in order to test them but not having any E.coli or yeast optimized signal peptide available at the registry is a critical obstacle in the project.
We would use Biobrick 2: BBa_K125310.
HisTag Options
Methionine + His Affinity Tag x 6: BBa_K133035 This part is only partially confirmed, so it would have to be tested.
We would put this 3 times in a row:
Sequence: atgcaccaccaccaccaccacatgcaccaccaccaccaccacatgcaccaccaccaccaccac
Note: It would take 3 ligation steps to obtain a larger peptide enriched Histidine, whereas in our main biobrick 1 and 3, it already comes in the same construct.
Terminator
There are several options for terminators, but we would use a double terminator in order to be sure that it works.
Double terminador: BBa_B0024
This sequence has a double terminator in several of reading frames with could be very useful.
Sequence:
aaataataaaaaagccggattaataatctggctttttatattctctctctagtatataaacgcagaaaggcccacccgaaggtgagccagtgtga
File:Bsas2012-Translate.png
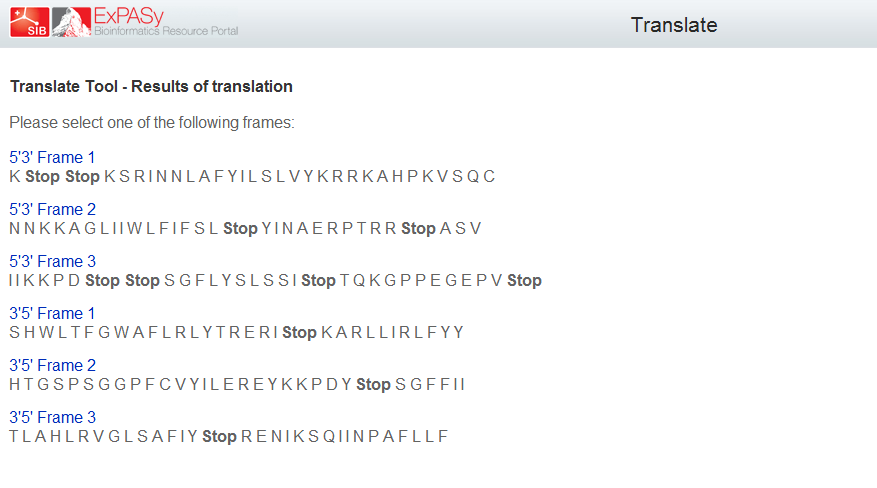
|
| Picture: General Design of Biobrick 5. |
 "
"