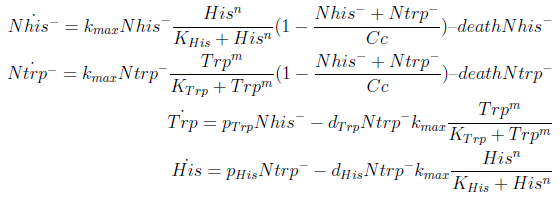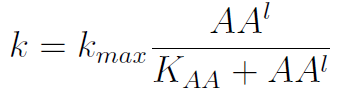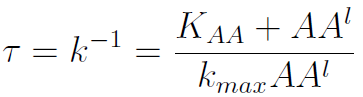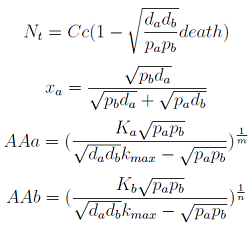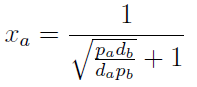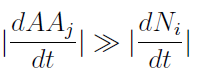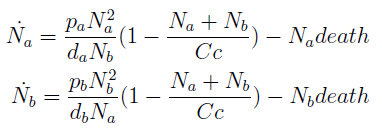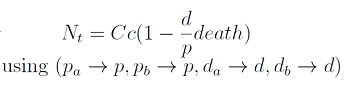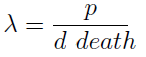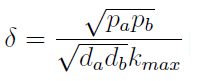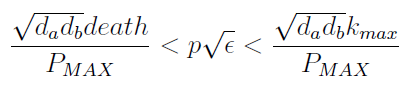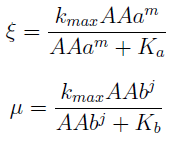Team:Buenos Aires/Project/Model
From 2012.igem.org
(→Variable Transformation) |
(→Variable Transformation) |
||
| Line 340: | Line 340: | ||
# Na exports more that could Nb possibly adsorb and AAa accumulates in the medium. For AAi >> Ki the ratio ξ -> 1. | # Na exports more that could Nb possibly adsorb and AAa accumulates in the medium. For AAi >> Ki the ratio ξ -> 1. | ||
| - | The AAs can be zero, so can the new variables. Rewriting the equation we see that each ratio is elevated to 2- (s+1)/s > 0 for s>1; so the variables are not really in the denominator. | + | * The AAs can be zero, so can the new variables. Rewriting the equation we see that each ratio is elevated to 2- (s+1)/s > 0 for s>1; so the variables are not really in the denominator. |
| - | χ stop growing (is constant) if and only if μ = ξ, n can’t equal 1 since n=1 doesn’t satisfy the DE for n. | + | |
| - | + | * χ stop growing (is constant) if and only if μ = ξ, n can’t equal 1 since n=1 doesn’t satisfy the DE for n. | |
| - | + | Here’s the great improvement over the old variables. The SS for χ, ξ and μ is independent of n; i.e. the conditions for which their DE equal zero. This means that the nullclines are independent as well. Now we have a reduction in the system's dimension. Same SS as 4x4 but this 3x3 system can be analyzed visually, | |
| - | + | * We can represent the vector field ''eq'' in space and track a trajectory that starts at (χ0, ξ0, μ0) | |
| + | |||
| + | * We plot the nullclines, see where they meet. | ||
| + | |||
| + | [[File:bsas2012-modeling-fig9.jpg]] | ||
| - | + | Figure 9. Nullclines | |
== Stability of the percentage in equilibrium == | == Stability of the percentage in equilibrium == | ||
Revision as of 04:54, 26 September 2012

Contents |
Modeling a synthetic ecology
To gain insight into the behavior of our crossfeeding design, to understand which parameters of the system are important, and to study the feasibility of the project, we decided to implement a model of the system. We found this interesting per se, as the model we need is that of a microbial community, an ecology.
The mathematical modeling of ecological system is an active field of research with a very rich and interesting history; from the works of Lotka and Volterra, who modeled predator-prey dynamics with ordinary differential equations, to Robert May’s chaotic logistic maps. In fact a lot of synthetic systems of interacting bacteria created in recent years were used to study these sort of models.
The crossfeeding model
In the crossfeeding design, each strain produces and releases to the medium an aminoacid the other strain(s) need to grow. The growth rate of each strain depends on the aminoacid (AA) concentration of those AA it can’t produce. In turn these concentrations depend on the abundance of the other strains. Therefore the growth of the strains is interdependent.
For simplicity and to be consistent with the experimental work, we decided to model two interacting strains, one that produces tryptophan (Trp) but requires histidine (His), and another that produces His but requires Trp.
We decided to use ordinary differential equations to model the system. This is a normal approach when studying population dynamics of this kind.
The model has four variables:
- [Nhis-] : the concentration of histidine dependent (Trp producing) yeast cells, in cells per ml
- [Ntrp-] : the total amount of tryptophan dependent (His producing) yeast cells, in cell per ml
- [his]: the concentration of histidine in the medium, in molecules per ml
- [trp]: the concentration of tryptophan in the medium, in molecules per ml.
To build the model we did the following assumptions:
- The growth rate of each strain depends on the concentration in the medium of the AA they can’t produce. As the concentration of this AA in the medium increases, so does the growth rate of the strain, until it settles at the growth rate observed in optimal conditions (doubling time of 90 min for yeast).
- There is a maximal density of cells the medium can support (carrying capacity)
- Each cell has a fixed probability of dying per time interval
- Each cell releases to the medium the AA it produces at a fixed rate
- Each strain only consumes the AA it doesn't produce
- The system reaches steady state.
In the equation for the temporal evolution of each population we take the growth rate as a Hill function of the amount of AA to which the strain is auxotrophic (it can't produce). The function captures the increase of the growth rate with the relevant AA concentration, until it reaches a plateau at the maximal growth rate of 90 minutes per cell division cycle (1/kmax).
K_AA is the effective concentration of AA at which half maximal growth rate is obtained.
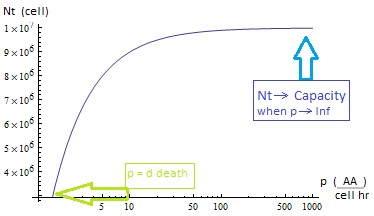
The second term accounts for other factors, not explicit in our model that can limit the concentration of cells in a given volume (carrying capacity) enabling the population to reach equilibrium. Given that our model relies on nutrient deficiency we can't rule out starvation so we added a term related to cell death.
The terms in the equations for the evolution of each [AA] in the medium are the fluxes of AA entering and leaving the medium. Each rate p is a measure of the AA produced and exported in the form of a protein by a cell. In our simulations it is defined as a fraction of the calculated maximum of elongation events.
The second term is the flux leaving the medium and entering the auxotrophic cells. We have assumed that the total amount of AA absorbed, d, during the cell cycle time τ is the required to build the daughter. One of the hypotheses built into our model is that τ will vary greatly with the concentration of nutrients available. We've used
The amount of AA entering the cells that produce it was considered negligible.
Steady State Solution
The 4 equations in [] were equaled to zero and the non-linear algebraic system solved using MATEMATICA to find their equilibrium values.
Here we change the notation a little to two generic strains a and b where population Na produces amino acid a, etc. We are interested in the value of the cell populations in equilibrium, more importantly in the fraction of each population. Thus a change was made to more convenient variables: the sum of both population and a strain's molar fraction.
{N_a ; N_b} → { N_t , x_a }
Only 3 fixed points were found for the system {N_t, X_a, AA_a, AA_b} (See Appendix) That means that for some solutions it’s not true that the four variables reach a steady state (SS) or equilibrium. There are initial conditions for which some of the variables will not stop growing!
- The first solution is clear. The culture will not thrive under certain conditions… to be determined!
- The second one has no physical relevance since it yields negative concentrations for the Amino Acids.
- The third one is the significant one. However the concentrations of Nt and AA can be negative; we must work within the region where is positive.
Regulation
Note that the percentage of each population in the community is a function of √((da pb)/(db pa)).
In particular
For positive values of d and p the range of the function is (0; 1), so far it’s consistent with what we expect.
The percentages of the strains in the culture in equilibrium are regulated by the production and export of AA – represented in the model through pa and pb; independent of initial conditions!!
In fact we need only know the ratio between these two parameters to control it:
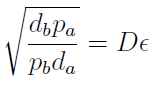 (5)
(5)
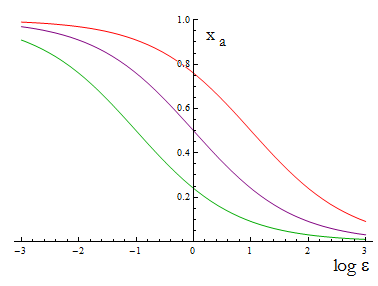
The next figure illustrates how the mole fraction x_a varies with the variable ε.
Figure 1. Mole fraction x_a as a fucntion of the ratio of protein export for values of D=0.1,1,10.
The ratio D is fixed for two A.A., this determines the range of percentages of each strain accessible for any selection of biobricks.
Parameters
The parameter estimation process is shown in xx section(linked)
| Value | Units | |
| kmax | 0.4261 | 1/hr |
| K his | 2.588e16 | AA/ml |
| K trp | 1.041e16 | AA/ml |
| n his | 1.243 | |
| n trp | 1.636 | |
| Cc | 3.0 10^7 | cell/ml |
| death | 3 to 7 | days |
| p his | p 2.16e8 | AA/ cell hr |
| p trp | p ε 2.16e8 | AA/ cell hr |
| d his | 6.348e8 | AA/cell |
| d trp | 2.630e7 | AA/cell |
Table 1.
Model Analysis
The standard procedure when dealing with a non-linear system is to find the fixed points(FP) and later analyze their stability; same as with a linear system (**). The goal is to determine all possible trajectories, i.e. for any initial conditions how do the variables evolve?
How is this behavior governed by the model’s parameters?
How do the trajectories change as the parameters vary? Does any new FP or close orbit appear?
There was a major difficulty in the analysis of the model because of the dimension of the problem - 4 variables. The analytical results yielded by the usual tools (nullclines, linearization) could not be simplified so that all the relevant properties of the system could be understood in terms of the parameters.
We obtained the 4x4 Jacobian - evaluated in the FP - and later its eigenvalues. The expression found for them, a function of the relevant parameters, is too long and can’t be simplified in a way that reveals changes in stability, etc.
Besides that, we couldn’t do a visual analysis of the nullclines to have a qualitative idea of what goes on.
A very common way to proceed in such a case is to reduce the model, ideally to a two-dimensional system. Sometimes one of the variables varies so slowly or that much faster than the ones we are interested in, that it can be considered constant when studying the dynamics of the rest. This is a standard procedure in the study of chemical reactions called Quasi Steady State Approximation.
- QSSA: Knowing that initially the variation of AA in the medium is much faster than the yeast‘s populations.
We supposed that it reaches SS before the other. Therefore taking dAAj/dt=0 we can rewrite the AminoAcids as a function of (Na,Nb) in a reduced 2x2 model for the evolution of the populations.
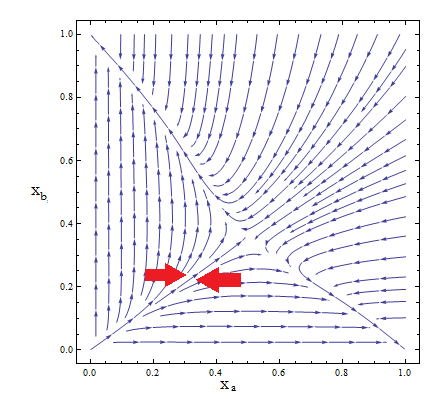
Thus a phase portrait for the nullclines and trajectories was drawn.
Figure 2
Although the reduced model presents the same fixed point, it’s a saddle point.
What does that mean? Pick any point for the plane; those are your initial conditions for the concentration of each population. Now to see how the community evolves with time follow the arrows. No matter how close to the FP we start, the community is overtaken by one of the two strains.
- The stable FP are {Na-> Nt, Nb-> 0} and {Na-> 0, b->Nt}.
- Only by taking i.c. along the stable manifold –direction marked between the red arrows for clarification in Figure 2 – do we reach the FP we wanted as t-> inf.
This is not a representation of auxotrophic community!
Indeed a linearization of the set of ODEs around the FP yields instability regardless of the parameters. One of them is always positive.
{2death,death (1-√((pb pa)/(db da))/death)}
There is no agreement between the predictions found here and the simulations; we won’t pursue this reduction any further.
- We also noticed that the Hill coefficients are similar to one. So we explored the possibility of approximating them to one; is there a relevant change or not?
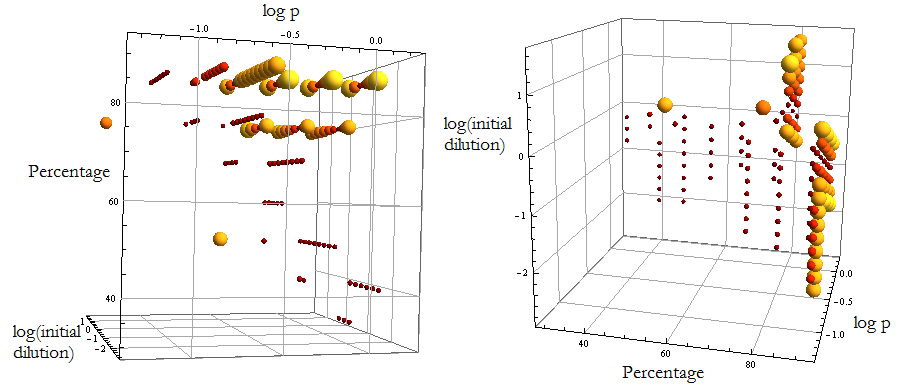
Simulations
We’ve relied on numerical simulations to explore possible behaviors.
To run the simulations all that is left is to choose values {p, ε, i.c.}.
Since this is merely a framewok we have taken values p from a log scale from -3 to 1.
ε was ranged from 0.0001 to 4; which varies the fraction of each population from 10% to 90%.
Cells’ initial concentration: dilutions from 1/10000 to 1/100 of the capacity (Cc).
AAs’ initial concentration: null.
Die or thrive?
Some basic properties were observed by simplifying the system; we wanted to check that our model was sound.
Figure 3.
The condition for culture growth is λ > 1. This ratio λ between two rates determines whether the system is able to produce and absorbed the nutrients needed for the cell cycle before its death. If it doesn’t hold the culture dies, ie the model predicts a negative amount of cells; which doesn’t make sense.
The other condition is related to extracellular AAs. Let’s define
The concentration of extracellular AAs is positive only if δ < 1. In fact as δ -> 1the concentration -> infinity because δ = 1 is a vertical asymptote. This ratio δ between two rates is related to our proposed regulation; if the amount of nutrients produced exceeds the required one for a 90 minutes cycle, the medium is saturated. Then the regulation fails.
Parameter Space and Solutions
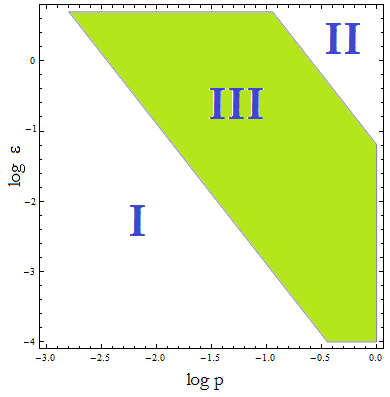
Uniting the conditions for λ and δ we see that p and &epsilon must be bound for our solution to make sense:
Figure 4. Regions in the parameter space that present different types of solutions. Only Region III both conditions are true.
Now, let’s classify these regions numerically to see whether the ideas we just presented are supported. Taking random values from each region we found the following:
I . The culture doesn’t grow, it decays with varying velocities for any i.c.. This is consistent with solution 1, where NT = 0.
II. The extracellular AA keep growing and the medium is saturated. The culture reaches equilibrium for every i.c. no matter how low.
The growth rate tends to the theoretical 90 min and the AA dependence disappears from the equations for each population. Remarkably the mole fraction is still independent of initial concentration of cells.
III. The four variables reach SS. The mole fraction is solely dependent on ε according to the formula on eq [].
The total population of the community (Nt) is the same as in Region II.
This is the region where the auxotrophy leads to regulation of the community.
However the culture doesn’t grow for all i.c. – will get back to this point shortly.
We had postulated that the solution doesn’t hold in II and expected a dramatic change in the system’s behavior. However there is a smooth transition between II and III. For a fixed ε we can step from III into II by increasing ‘’p’’. Once we cross the threshold we notice that the total population of the community does not vary from one region to the next and though the AA continues to grow over time (and accumulate) it does so slowly (compare to what is expected for a vertical asymptote). More so, the formula [] is a good approximation for the mole fraction in the immediate area; the difference between the two increases gradually.
There’s no pointed change caused by the asymptote, in fact the two regions can’t be distinguished experimentally; so small is the difference between the mole fractions of a strain in each side of the frontier.
An important test: under which initial conditions do the populations thrive or die has to be understood in terms of Figure 4, not merely the conditions associated with Figure 3.
We noticed that the culture’s survival in region III is also dependent on the Ks even though is not explicit in ay e formula so far. Rather for a fixed value of K there is a i.c. threshold below which the culture doesn’t prosper.
This is reasonable considering that it introduces another time scale. Not only have the cells to produce and absorb the required amounts of AA before they die, their ability to grow is restricted by the concentration of AA in the medium. The cells must produce enough amino acids so that [AA] is of the order of K in the same time frame to insure their prosperity.
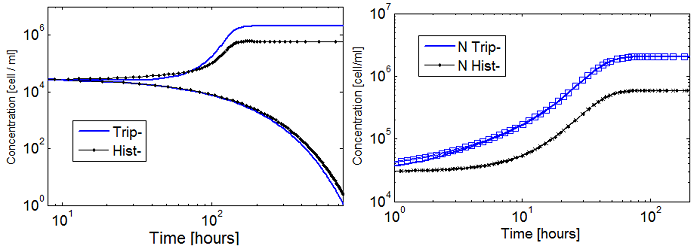
Why is this important? Originally we took both Hill coefficients equal to one, is close enough, right?
Well, the whole system didn’t seem viable; it took over 100 hours for the fastest cultures to grow. Now using the fitted coefficients; the time scale becomes reasonable again.
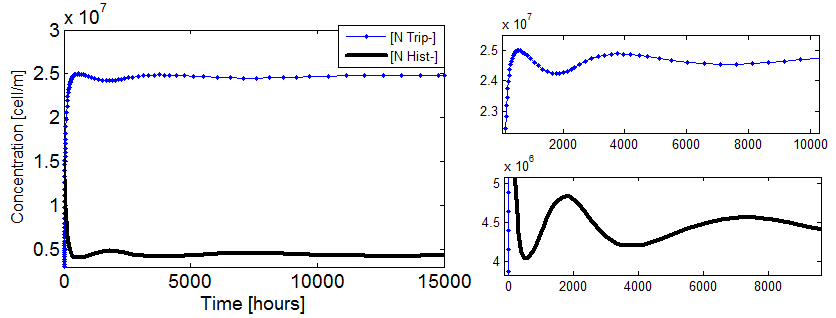
Figure 5. Evolution in time of two cultures that only differ in the i.c. for n_ hill =1 (left)and n_hill >1 (right).
When n_ hill =1 only the one with higher i.c. reaches a non-trivial steady state (ss). The other slowly dies. However for n_ hill =1.5 there is no clear difference between the cases with high and low i.c.. Note that it reaches SS faster as well.
The survival of an auxotrophic community is dependent on both the apparent dissociation constants (Ks) and the Hill coefficient as shown clearly on Figure 5. There are two effects here:
Shorter time scale to SS.
Larger subspace of i.c. that lead to SS.
So when creating an auxotrophy, pick a nutrient with an appropriate Hill coefficient and K. Check the range and stability of your model estimating the values of the rest of the parameters.
Our case is similar to the one shown in figure 5 with both hill coefficients > 1. Region III is less sensitive to i.c.
Relaxation time
Not only are we interested in predicting the fraction of each strain in the culture, we want it within a time frame.
Let’s call Τ the time it takes one of the populations to reach 90% of its SS value (relaxation time). We rely again on numerical simulations to obtain those set of parameters where Τ is less than 65 hours, for example.
Creating a mesh we explored the parameter space (p, ε) for different initial conditions to visualize when the model’s prediction and the numerical result of the simulations differ in an amount lower than the given error; before 65 hours. We won’t limit the mesh to Region III only.
We’ve taken 5% of the value as error; considering it an accurate estimation of an experimental measure’s precision.
Figure 6.
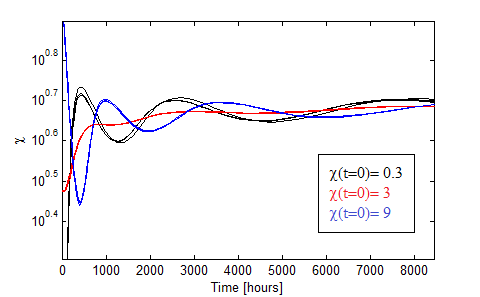
Oscillations
One interesting effect appears if the initial concentration of AA in the medium are of the order of the Ks. Damp oscillations are observed!!
Figure 7.
Variable Transformation
Let’s review what we’ve learned so far from our system.
- Seems stable,
- capable of oscillations,
- there are two cases with a stable population and a defined mole fraction for each strain, one where the AAs are in SS and one where they aren’t.
- The set of i.c. for which the culture thrives is dependent on the Ks.
Yet all this information is not so trivial to find – especially the oscillations discovery was a fluke. There had to be a transformation where all these properties were more accessible or evident.
What we did so far was to write the results in more convenient variables.
Now we will try to find a transformation to a new set of variables, whose evolution is simpler to study and write a new set of ODEs for them.
For instance the AAs only appear as the argument of Hill functions, so why not just work with those rations. We defined:
This is better because now ξ is constant when:
- AAa reachs steady state.
- Na exports more that could Nb possibly adsorb and AAa accumulates in the medium. For AAi >> Ki the ratio ξ -> 1.
- The AAs can be zero, so can the new variables. Rewriting the equation we see that each ratio is elevated to 2- (s+1)/s > 0 for s>1; so the variables are not really in the denominator.
- χ stop growing (is constant) if and only if μ = ξ, n can’t equal 1 since n=1 doesn’t satisfy the DE for n.
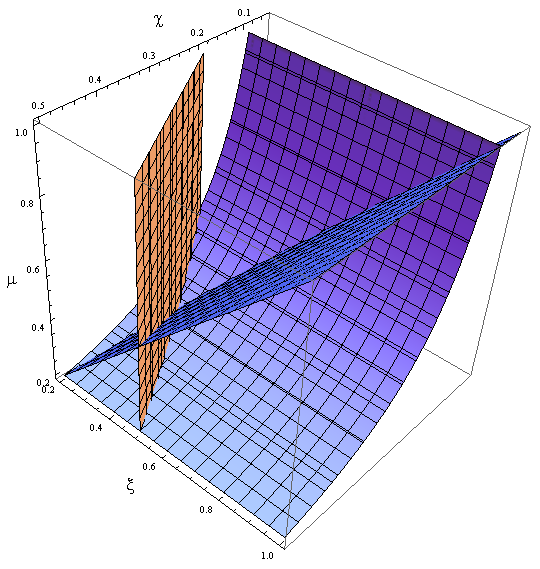
Here’s the great improvement over the old variables. The SS for χ, ξ and μ is independent of n; i.e. the conditions for which their DE equal zero. This means that the nullclines are independent as well. Now we have a reduction in the system's dimension. Same SS as 4x4 but this 3x3 system can be analyzed visually,
- We can represent the vector field eq in space and track a trajectory that starts at (χ0, ξ0, μ0)
- We plot the nullclines, see where they meet.
Figure 9. Nullclines
Stability of the percentage in equilibrium
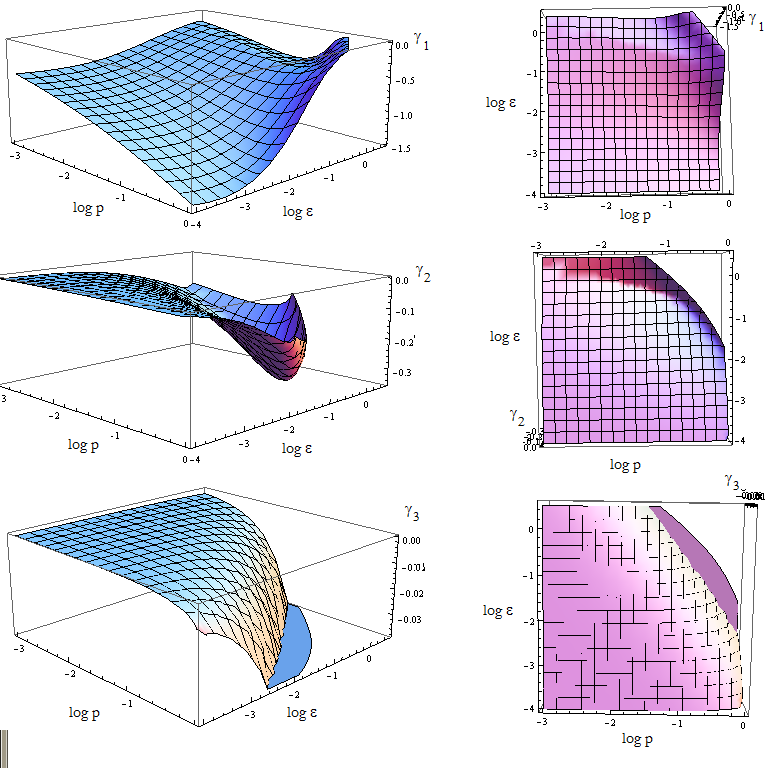
Let’s go the linearization of our 3x3 set of ODEs close to the FP. We said that n wasn’t relevant to the location of the fixed point, but now we have to consider it while examining the eigenvalues.
Once again the expresion for the three eigenvalues γ is very long. We replace the mayority of the parameters with the value we estimated, and n for its SS value - this is not always the case.
We are in condition to study the stability of the FP when we vary p_a and p_b. The results are shown vs variables p and ε in figure 10.
The FP is not stable if al least one of the eigenvalues is positive.
Figure 10.
Since the three eigenvalues are negative, it's a stable node for this set of parameters. There are however some points for which there's no value plotted. Taking (p, ε) from that region we obtain complex eigenvalues.
Parameter selection
We've estimatated values for all the parameter in the model to check is viability.
- The Ks found in the equations are a measure of the concentration of A.A. in the medium required to reach half the maximum rate of growth. Curves OD vs [AA] were measured experimentally for each amino acid and the data fitted to a Hill equation using MATLAB’s TOOLBOX: Curve Fitting Tool.
Figure aux his and aux trp.
The values were taken from the fits are
His- :
K_dil = 0.3279
ODmax = 62.53
n = 1.243
R-square: 0.9376
Trp- :
K_dil = 0.1770
ODmax = 57.46
n = 1.636
R-square: 0.9905
The results were then converted to the units chosen for the simulations.
K = K_dil * [AA 1x] # Avog / (Molar mass AA), where [AA 1x] = 0.02 mg/ml
- K trp= 0.1770 * 5.88e16 AA / ml.
- K his= 0.3279 * 7.8e16 AA / ml.
- The capacity of the system taken in the model is the one often used in the lab that works with these yeast strains:
- Cc= 3e7 #cell/ml.
- The death rate was taken between 3 and 7 days.
- The rate of “production and export” was given a top value (P_MAX) 1% of the maximum estimated number of elongation events for a yeast cell per hour. Our model does not explicitly include time as a variable, that means that the production is switched ON all the time.
Elongation events (peptidil transferase reactions): 6e6 1 / cell sec [ref]
P_MAX = 2.16e8 1/ cell hour
These parameters will be regulated experimentally (ε , p) to alter the fraction between populations.
- p trp = p* ε*2.16e8 aa / cell hour
- p his = p*2.16e8 aa / cell hour
where p < 1 controls the fraction of the top value and ε the ratio between the export of each amino acid.
- d is the total number of amino acids in a yeast cell –same as the ones needed to create a daughter -per cell:
d = # A.A. per cell = (mass of protein per yeast cell [ref]) * relative abundance of AA * # Avog / AA's molar mass.
- d trp= 2.630e7 AA / cell
- d his= 6.348e8 AA / cell
Appendix
solutions model1
Sol2
Sol3
 "
"