Team:British Columbia/Desulfurization
From 2012.igem.org

Synthetic Syntrophy
The field of synthetic biology has seen the development of many biological monocultures capable of performing a wide range of novel functions. In contrast to this current paradigm, microbes have naturally evolved to survive as members of dynamic communities with distributed metabolism. This “divide and conquer” strategy allows the community to perform more complicated metabolic processing than would be possible in single microorganisms while being resilient to environmental changes. Despite very recent proof of concepts in developing model microbial consortia, or synthetic ecology, questions remain as to whether complex metabolic pathways can be engineered in context of microbial populations. The 2012 University of British Columbia iGEM team sets a precedent by engineering a tunable consortium with a distributed 4S desulfurization pathway for increased efficiency in the removal of organosulfurs in heavy oils and bitumen resources.</br></br>
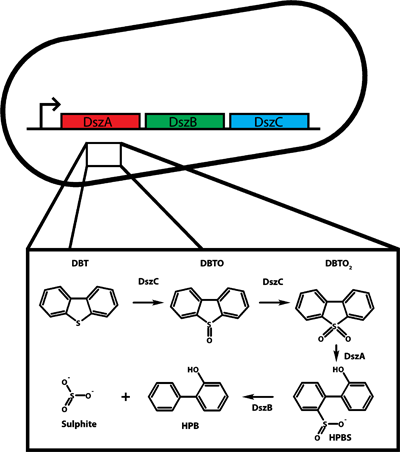
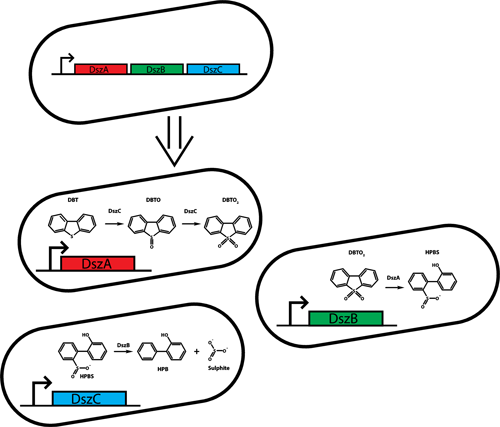
To create a distributed metabolic network, the three catalytic genes of the Dsz operon were separated into the three E. coli strains developed for our tunable consortium. As stated in the consortia objective, this is extremely important as it will allow us to refine and optimize the metabolic potential of the consortium.
Why the Dsz operon?
We choose the Dsz operon for a number of reasons. As stated above, bio-desulphurization has the potential to help relieve serious environmental problem associated with DBT content of fossil fuels. However, the Dsz operon also has a number of advantages directly related to our consortium. First, the pathway has previously been expressed functionally in E. coli, allowing us to obtain a working copy of the pathway expressed in an E. coli monoculture (REF). This will let us directly compare the efficiency of the single cell pathway with our distributed metabolic network. Also, the availability of a working pathway in e.coli indicates that all of the required enzymes will be active when recombinantly expressed in our e. coli consortium strains. Finally, the desulphurization activity of the 4S pathway can be easily monitored and quantitated by HPLC analysis (REF).</br>
 "
"