Team:Bielefeld-Germany/Labjournal
From 2012.igem.org
| Line 43: | Line 43: | ||
<!-- tab site --> | <!-- tab site --> | ||
<div id="labsite"> | <div id="labsite"> | ||
| - | |||
<div><h3>Prologue</h3> | <div><h3>Prologue</h3> | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
</html> | </html> | ||
| Line 103: | Line 99: | ||
<html> | <html> | ||
| + | |||
<table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | ||
<ul> | <ul> | ||
| - | <li class="toclevel-1 tocsection-1"><a href="# | + | <li class="toclevel-1 tocsection-1"><a href="#Week_1_.2804.2F30_-_05.2F06.2F12.29"><span class="tocnumber">1</span> <span class="toctext">Week 1 (04/30 - 05/06/12)</span></a> |
<ul> | <ul> | ||
<li class="toclevel-2 tocsection-2"><a href="#Monday_April_30th"><span class="tocnumber">1.1</span> <span class="toctext">Monday April 30th</span></a></li> | <li class="toclevel-2 tocsection-2"><a href="#Monday_April_30th"><span class="tocnumber">1.1</span> <span class="toctext">Monday April 30th</span></a></li> | ||
| - | |||
| - | <li class="toclevel-2 tocsection-4"><a href="# | + | <li class="toclevel-2 tocsection-3"><a href="#Tuesday_May_1th"><span class="tocnumber">1.2</span> <span class="toctext">Tuesday May 1th</span></a></li> |
| - | <li class="toclevel-2 tocsection-5"><a href="# | + | <li class="toclevel-2 tocsection-4"><a href="#Wednesday_May_2th"><span class="tocnumber">1.3</span> <span class="toctext">Wednesday May 2th</span></a></li> |
| - | <li class="toclevel-2 tocsection-6"><a href="# | + | <li class="toclevel-2 tocsection-5"><a href="#Thursday_May_3th"><span class="tocnumber">1.4</span> <span class="toctext">Thursday May 3th</span></a></li> |
| + | <li class="toclevel-2 tocsection-6"><a href="#Friday_May_4th"><span class="tocnumber">1.5</span> <span class="toctext">Friday May 4th</span></a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 196: | Line 193: | ||
'''Team Bacterial Laccases''': We did Colony PCR on the transformed the ''Bacillus pumilus'' CotA plasmid. Unfortunately the control with colony PCR didn't work. So we just picked some colonies for plasmid isolation in the hope that on the AMP plate were no false positives colonies. | '''Team Bacterial Laccases''': We did Colony PCR on the transformed the ''Bacillus pumilus'' CotA plasmid. Unfortunately the control with colony PCR didn't work. So we just picked some colonies for plasmid isolation in the hope that on the AMP plate were no false positives colonies. | ||
<html> | <html> | ||
| - | |||
</p> | </p> | ||
| + | </div> | ||
| - | |||
| - | |||
<div><h3>Summary of Week 2</h3> | <div><h3>Summary of Week 2</h3> | ||
<p class="more"> | <p class="more"> | ||
| Line 229: | Line 224: | ||
</ul> | </ul> | ||
</td></tr></table> | </td></tr></table> | ||
| - | |||
</html> | </html> | ||
| Line 283: | Line 277: | ||
* '''Team Activity Tests''': For some pre test and characterization for our future laccase activity standard we ordered [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE laccase] from ''Trametes versicolor''. As well we had to order a substrate that the laccase could use to demonstrate its abilities. According to the literature [http://www.sigmaaldrich.com/catalog/product/sigma/a1888?lang=de®ion=DE ABTS] is a well working substrate to characterize oxidizing enzym activity. So we ordered. | * '''Team Activity Tests''': For some pre test and characterization for our future laccase activity standard we ordered [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE laccase] from ''Trametes versicolor''. As well we had to order a substrate that the laccase could use to demonstrate its abilities. According to the literature [http://www.sigmaaldrich.com/catalog/product/sigma/a1888?lang=de®ion=DE ABTS] is a well working substrate to characterize oxidizing enzym activity. So we ordered. | ||
| - | <html> | + | <html> |
| - | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 297: | Line 290: | ||
<p> | <p> | ||
</html> | </html> | ||
| - | + | ==Week 3 (05/14 - 05/20/12)== | |
| + | |||
| + | __NOTOC__ | ||
| + | <html> | ||
| + | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | ||
| + | <ul> | ||
| + | <li class="toclevel-1 tocsection-1"><a href="#Week_3_.2805.2F14_-_05.2F20.2F12.29"><span class="tocnumber">1</span> <span class="toctext">Week 3 (05/14 - 05/20/12)</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-2 tocsection-2"><a href="#Monday_May_14th"><span class="tocnumber">1.1</span> <span class="toctext">Monday May 14th</span></a></li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-3"><a href="#Tuesday_May_15th"><span class="tocnumber">1.2</span> <span class="toctext">Tuesday May 15th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-4"><a href="#Wednesday_May_16th"><span class="tocnumber">1.3</span> <span class="toctext">Wednesday May 16th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-5"><a href="#Thursday_May_17th"><span class="tocnumber">1.4</span> <span class="toctext">Thursday May 17th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-6"><a href="#Friday_May_18th"><span class="tocnumber">1.5</span> <span class="toctext">Friday May 18th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-7"><a href="#Saturday_May_19th"><span class="tocnumber">1.6</span> <span class="toctext">Saturday May 19th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-8"><a href="#Sunday_May_20th"><span class="tocnumber">1.7</span> <span class="toctext">Sunday May 20th</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </td></tr></table> | ||
| + | </html> | ||
| + | |||
| + | ''' weekly seminar:''' | ||
| + | * first lab service: Robert | ||
| + | * our GFP, which we wanted to use for the summer school for pupils, does not work | ||
| + | * first competent cells have to be made: Julia S. and Robert | ||
| + | * Decision to buy a commercial laccase to establish the analytics and the enzyme tests | ||
| + | * our expose has to be translated into english: Malak | ||
| + | * last planning for our waver sell | ||
| + | * Julia S. is creating a vector for ''Pichia pastoris'' and is now looking for sequences | ||
| + | * The [http://www.bmbf.de/en/index.php BMBF] invites all german iGEM teams to Berlin to attend at the Biotechnologie2020+ strategy process | ||
| + | |||
| + | |||
| + | ===Monday May 14th=== | ||
| + | ===Tuesday May 15th=== | ||
| + | |||
| + | ===Wednesday May 16th=== | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | :*For cloning our laccases we need pSB1C3 backbone. Therefore we we transformed [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (pSB1C3 with RFP) in competent KRX cells. | ||
| + | |||
| + | ===Thursday May 17th=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': Plasmid isolation of <partinfo>BBa_J04450</partinfo>. | ||
| + | |||
| + | * '''Team Modeling''': | ||
| + | **Meeting Mrs. Lutter, a mathematics prof. of our course of studies and looking for our first model of a metabolic pathway, finding out, that we don't need such a complex model. Start thinking that we want and what we need. | ||
| + | |||
| + | ===Friday May 18th=== | ||
| + | * '''Team Activity Tests''': Our ''T.versicolor'' laccase and the ABTS arrived! We couldn´t wait to start, so we set up the stock solutions we will need, such as sodium acetat buffer (pH 5), 10 mM ABTS and deluted laccase. | ||
| + | |||
| + | ===Saturday May 19th=== | ||
| + | ===Sunday May 20th=== | ||
<html> | <html> | ||
</p> | </p> | ||
| Line 304: | Line 348: | ||
| - | <div><h3>Summary of Week | + | <div><h3>Summary of Week 4</h3> |
<p class="more"> | <p class="more"> | ||
</html> | </html> | ||
| Line 312: | Line 356: | ||
<p> | <p> | ||
</html> | </html> | ||
| - | + | ==Week 4 (05/21 - 05/27/12)== | |
<html> | <html> | ||
| - | </ | + | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> |
| - | + | <ul> | |
| + | <li class="toclevel-1 tocsection-1"><a href="#Week_4_.2805.2F21_-_05.2F27.2F12.29"><span class="tocnumber">1</span> <span class="toctext">Week 4 (05/21 - 05/27/12)</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-2 tocsection-2"><a href="#Monday_May_21st"><span class="tocnumber">1.1</span> <span class="toctext">Monday May 21st</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-3"><a href="#Tuesday_May_22nd"><span class="tocnumber">1.2</span> <span class="toctext">Tuesday May 22nd</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-4"><a href="#Wednesday_May_23rd"><span class="tocnumber">1.3</span> <span class="toctext">Wednesday May 23rd</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-5"><a href="#Thursday_May_24th"><span class="tocnumber">1.4</span> <span class="toctext">Thursday May 24th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-6"><a href="#Friday_May_25th"><span class="tocnumber">1.5</span> <span class="toctext">Friday May 25th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-7"><a href="#Saturday_May_26th"><span class="tocnumber">1.6</span> <span class="toctext">Saturday May 26th</span></a></li> | ||
| - | + | <li class="toclevel-2 tocsection-8"><a href="#Sunday_May_27th"><span class="tocnumber">1.7</span> <span class="toctext">Sunday May 27th</span></a></li> | |
| - | + | </ul> | |
| + | </li> | ||
| + | </ul> | ||
| + | </td></tr></table> | ||
</html> | </html> | ||
| - | + | ||
| - | < | + | ''' weekly seminar:''' |
| - | + | * Lab service: Isabel | |
| - | < | + | * We try to establish a collaboration with the iGEM team from [https://2012.igem.org/Team:SDU-Denmark SDU-Denmark] |
| - | </ | + | * Got our distribution kits |
| - | + | * First successful cloning and cultivations | |
| + | * Who wants to be a summer school teacher? | ||
| + | * We will not travel to the ACHEMA because only local teams are invited | ||
| + | * Do we want to participate in the Biolympics? (It's a sports party with fun organized by the [http://bts-ev.de/ bts]) | ||
| + | |||
| + | |||
| + | ===Monday May 21st=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** We wanted to clone our laccase PCR products xccl and ecol in pSB1C3 backbone. Therefore we did some restriction digests on the PCR products and the vector [http://partsregistry.org/Part:BBa_J04450 BBa_J04450]. | ||
| + | |||
| + | * '''Team Modeling''': Our aims for modeling: | ||
| + | ** model the expression of the laccases in the organisms. | ||
| + | ** model the activity of our enzymes. | ||
| + | ** model the interesting parts of a clarification plant (the part witch are interesting for our cleaner. | ||
| + | |||
| + | ===Tuesday May 22nd=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | :*Ligation of the digested PCR products in pSB1C3 backbone and transformation in KRX electro-competent cells. | ||
| + | |||
| + | ===Wednesday May 23rd=== | ||
| + | *''' Team Student Academy:''' | ||
| + | ** Repetition of the transformation of [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 BBa_I13522] with new competent ''E. coli'' KRX cells. Got intense red fluorescing colonies but no green fluorescing colonies. Made a backup of ''E. coli'' KRX [http://partsregistry.org/Part:BBa_J04450 BBa_J04450]. | ||
| + | ** Asking for other plasmids containing GFP at the working groups of our University. | ||
| + | |||
| + | *'''Team Cloning of Bacterial Laccases''': | ||
| + | ** After there were no colonies on our pSB1C3 + xccl(T7)_His (''Xanthomonas campestris'') transformation plate we did the transformation with the same ligation preparation again. The other ligation with pSB1C3 + ecol(T7)_His (''E. coli'') showed colonies so we started colony PCRs to find positive colonies. Sadly the colony PCRs showed no products but the problem was that we just had the long overhang primers (E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS ). Therefore we ordered the F and R [https://2012.igem.org/Team:Bielefeld-Germany/Protocols#Primers primers]. | ||
| + | |||
| + | ===Thursday May 24th=== | ||
| + | *'''Team Student Academy:''' | ||
| + | ** Made a liquid culture of ''E. coli'' KRX with [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] at 30 °C. There was no fluorescence. | ||
| + | ** Transformation of [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] into ''E. coli'' KRX. Also got intense red fluorescing colonies. | ||
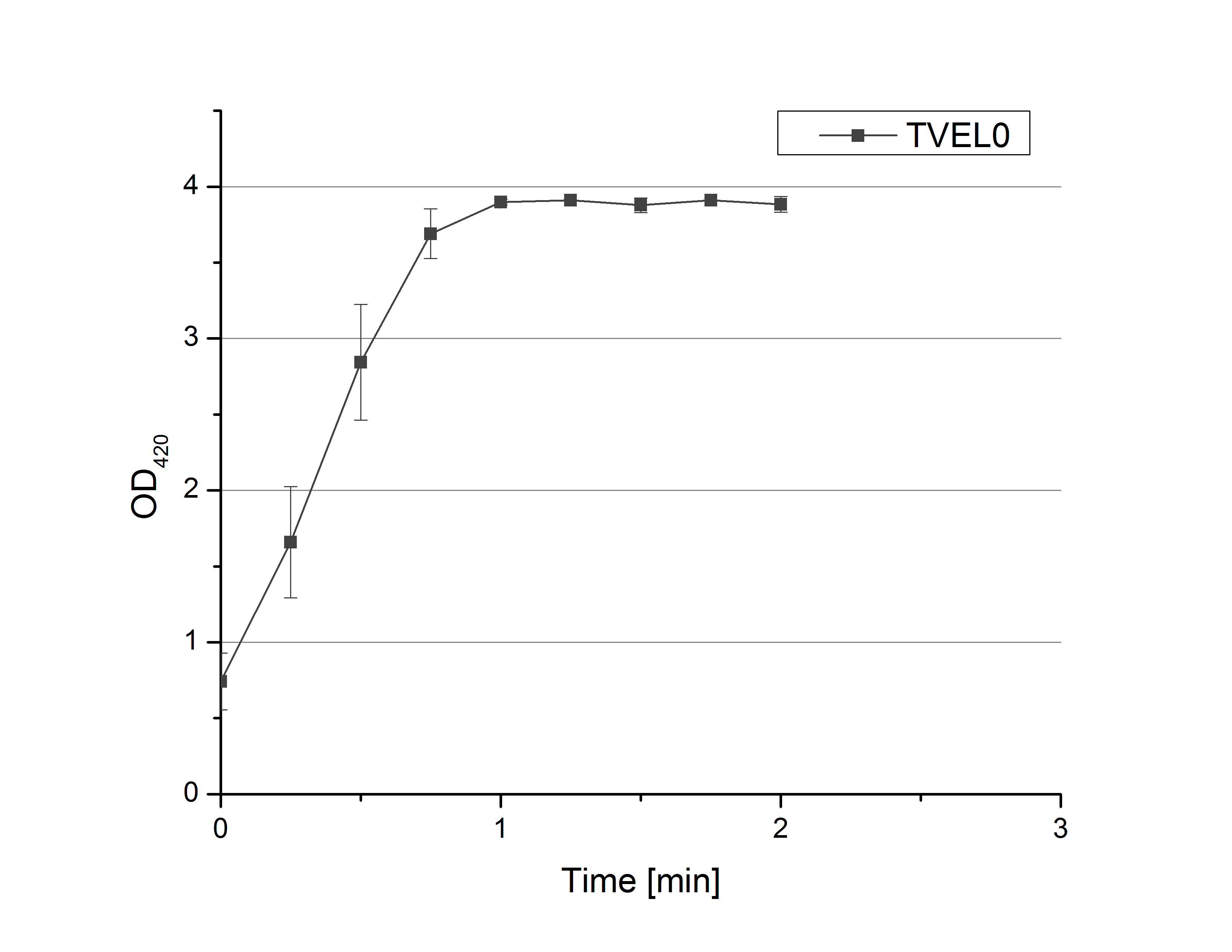
| + | [[File:Bielefeld2012_initialGraph_TVEL0activity.png|200px|thumb|right|Fig. 1: Oxidation of ABTS by TVEL0 reporting the activity of TVEL0 (n=4). Measurement setup see text.]] | ||
| + | *'''Team Activity Tests:''' | ||
| + | **On our schedule today was testing our bought laccases and improving our protocol in case it won't work. We got familiar with the flat bottom microplates and got a briefing into using the Tecan correctly. We started with the following setup: 100 mM sodium acetate (pH 5), 5 mM ABTS, 8 U TVEL0 laccase, ad 200 µL with deionized water. Then we measured the absorption at 420 nm every 15 seconds over a time period of 2 minutes. Immediately our samples turned dark blue but unfortunately the changeover was out of range to be detected by Tecan. With this information we needed to reduce the laccase concentration to get measurable results. We have chosen to try another measurement with 0.1 U TVEL0 laccase and it worked! The result was a nice saturation curve but it reached a too high, because not good measurable, optimum within 1 minute (Fig. 1). To avoid the loss of important data in the beginning of the reaction and to reduce the saturation OD we decided to slow everything down by using less ABTS. | ||
| + | |||
| + | ===Friday May 25th=== | ||
| + | *'''Team Cloning of Bacterial Laccases''': | ||
| + | :* We had to do the PCR on ''T. thermo'' laccase again because after the purification of the last PCR product the DNA amount was very low. | ||
| + | *'''Team Student Academy:''' | ||
| + | ** Made a liquid culture of ''E. coli'' KRX with [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] at 30 °C. Result: There was no fluorescence. | ||
| + | |||
| + | ===Saturday May 26th=== | ||
| + | ===Sunday May 27th=== | ||
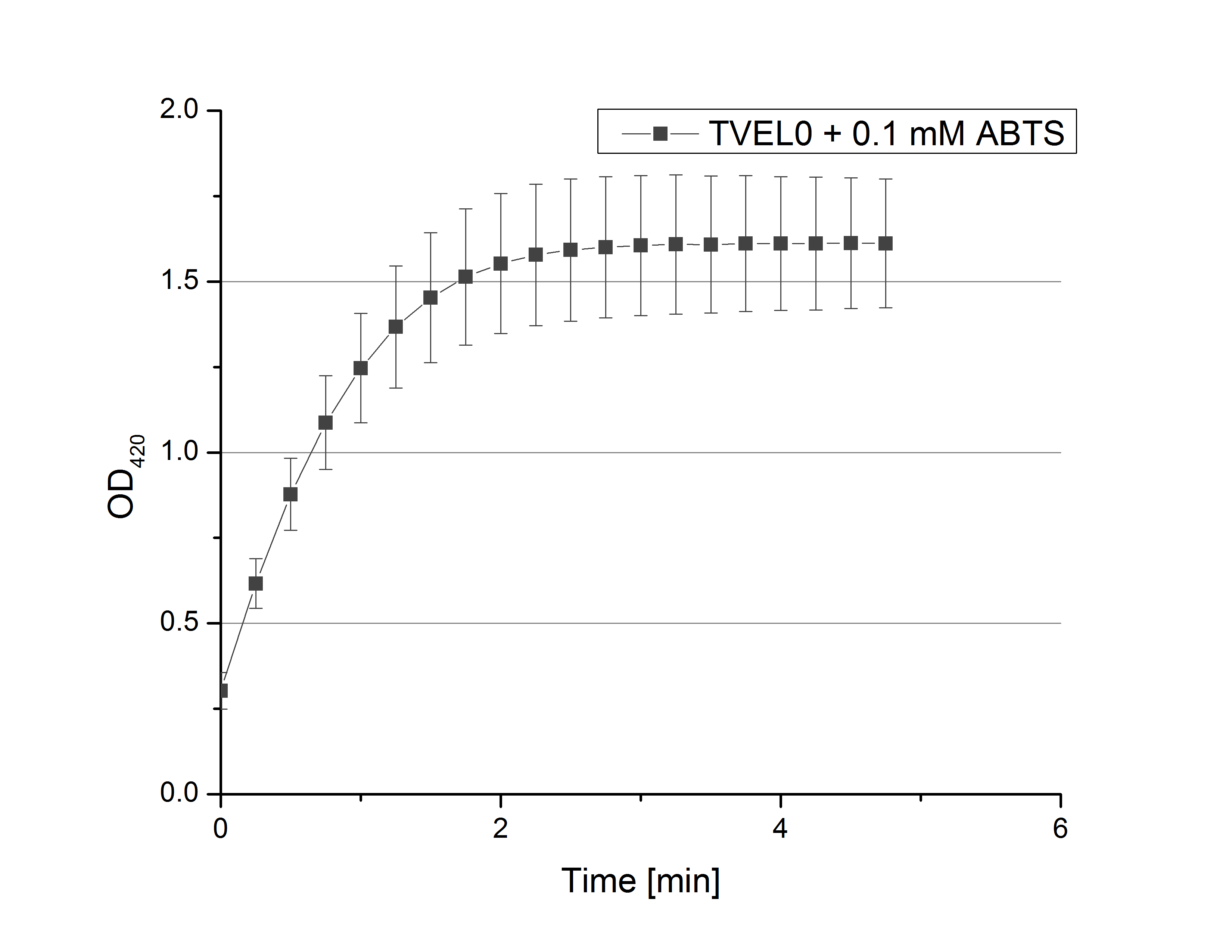
| + | [[File:Bielefeld2012_inititialActivity_0_1mM_ABTS.png|200px|thumb|right|Fig. 2: Oxidation of 0.01 mM ABTS by TVEL0 reporting the activity of TVEL0 (n=24).]] | ||
| + | *'''Team Activity Test:''' | ||
| + | **Remember our first activity measurements? The activity was awesome and reached quickly its saturation. So today we decided to have a lazy Sunday and reduce absorption maximum and reaction speed, too. For this we used 0.1 mM ABTS instead of 5 mM ABTS and we got good results. The saturation reached its maximum at a OD<sub>420</sub> of ~1.6 after roundabout 2 minutes (see Fig. 2). Taking together we halved the maximal OD<sub>420</sub> and doubled the reaction time until maximum is reached with using 0.1 mM ABTS instead of 5 mM ABTS. | ||
<html> | <html> | ||
</p> | </p> | ||
| Line 332: | Line 432: | ||
| - | <div><h3>Summary of Week | + | <div><h3>Summary of Week 5</h3> |
<p class="more"> | <p class="more"> | ||
</html> | </html> | ||
| Line 340: | Line 440: | ||
<p> | <p> | ||
</html> | </html> | ||
| - | + | ||
| + | ==Week 5 (05/28 - 06/03/12)== | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
| + | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | ||
| + | <ul> | ||
| + | <li class="toclevel-1 tocsection-1"><a href="#Week_5_.2805.2F28_-_06.2F03.2F12.29"><span class="tocnumber">1</span> <span class="toctext">Week 5 (05/28 - 06/03/12)</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-2 tocsection-2"><a href="#Monday_May_28th"><span class="tocnumber">1.1</span> <span class="toctext">Monday May 28th</span></a></li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-3"><a href="#Tuesday_May_29th"><span class="tocnumber">1.2</span> <span class="toctext">Tuesday May 29th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-4"><a href="#Wednesday_May_30th"><span class="tocnumber">1.3</span> <span class="toctext">Wednesday May 30th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-5"><a href="#Thursday_May_31st"><span class="tocnumber">1.4</span> <span class="toctext">Thursday May 31st</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-6"><a href="#Friday_June_1st"><span class="tocnumber">1.5</span> <span class="toctext">Friday June 1st</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-7"><a href="#Saturday_June_2nd"><span class="tocnumber">1.6</span> <span class="toctext">Saturday June 2nd</span></a></li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-8"><a href="#Sunday_June_3rd"><span class="tocnumber">1.7</span> <span class="toctext">Sunday June 3rd</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </td></tr></table> | ||
| + | |||
| + | ===Monday May 28th=== | ||
| + | * '''Team Student Academy:''' | ||
| + | ** Made liquid cultures of ''E. coli'' KRX with [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and with [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. Result: Intense red fluorescence. We made a glycerol stock and a plasmid isolation | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases:''' | ||
| + | **Digest of ecol(T7)_HIS, xccl(T7)_His and bpul(T7)_His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (pSB1C3 + RFP) with the same restriction enzymes, ligation of the digested products and transformation in ''E. coli'' KRX cells. | ||
| + | ** Since we have less bpul(T7)_His and tthl(T7)_His DNA, we set an PCR from the remaining PCR approach. | ||
| + | |||
| + | ===Tuesday May 29th=== | ||
| + | |||
| + | |||
| + | *'''Team Student Academy:''' | ||
| + | ** ''E. coli'' Mach1 with pMTE cp46 His received from the working group “Fermentation Engineering”, University Bielefeld. Plasmid contains genes for GFP and kanamycine resistance. We plated it and made a liquid culture at 37°C. Result: There was an intense fluorescence. We made a glycerol stock and a plasmid isolation. | ||
| + | |||
| + | *'''Team Cloning of Bacterial Laccases:''' | ||
| + | ** We did colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the ''E. coli'' laccase, the ''Xanthomonas campestris'' laccase and the ''B. pumilus'' laccase with the inducible T7 promoter and a His-tag. | ||
| + | |||
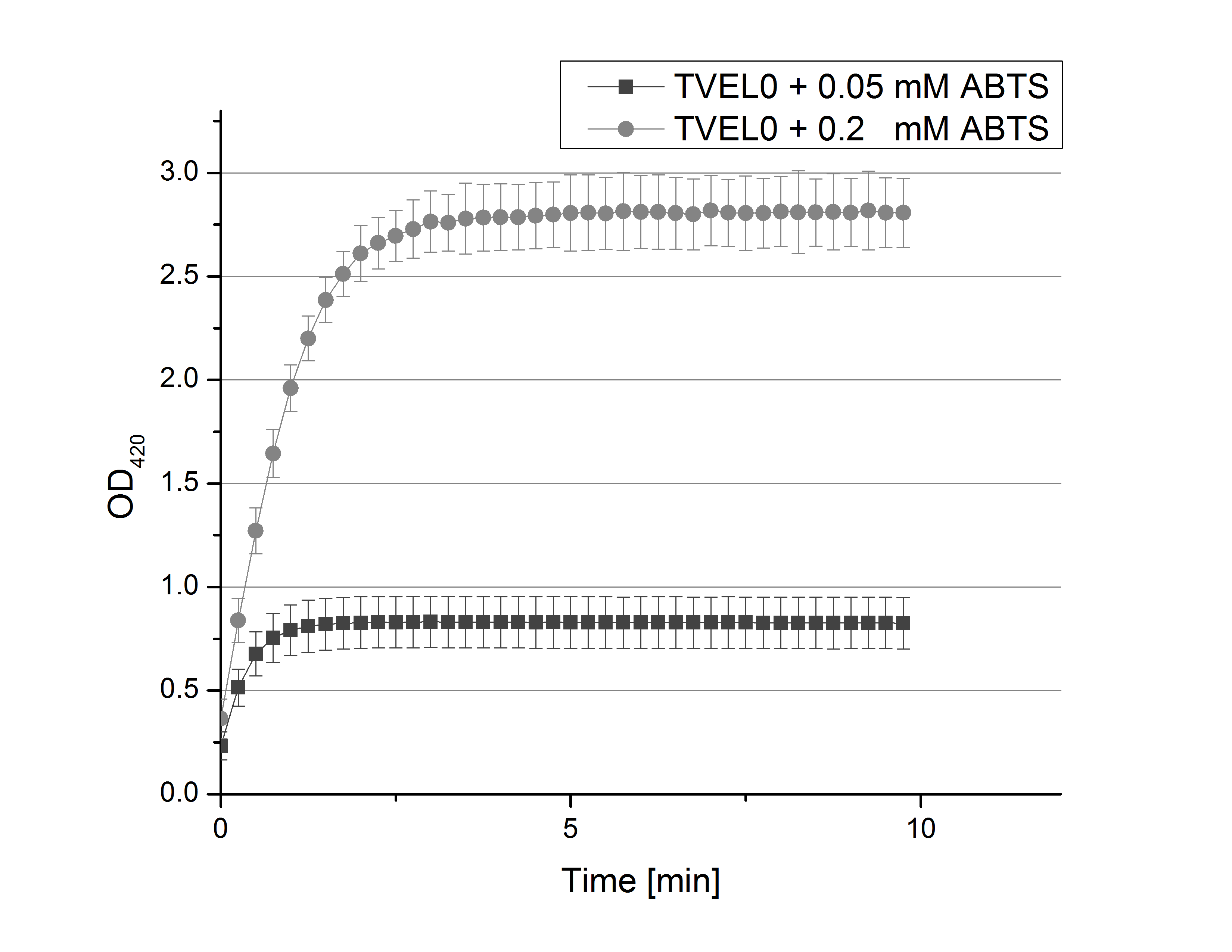
| + | [[File:Bielefeld2012_02_und_005_mM_ABTS.png|200px|thumb|right|Saturation curves of TVEL0 activity with 0.1 mM and 0.05 mM ABTS (n=24).]] | ||
| + | *'''Team Activity Tests:''' | ||
| + | **After establishing our recipe for activity measurements we were curious about different ABTS concentration and wanted to make sure we took the right one for our approach. With this in mind we did our activity measurement with 0.1 U TVEL0 laccase, 100 mM sodium acetate buffer (pH 5), ad 200 µl H<sub>2</sub>O dest., but with two new ABTS concentrations, namely 0.2 mM and 0.05 mM ABTS (see Fig. 1). It turned out that using 0.05 mM ABTS leads to a low maximum of saturation but reaches it quickly. With 0.2 mM ABTS the opposite occurs: the activity curve is saturated at a OD<sub>420</sub> of ~2.7 but it needs more time to reach its maximum. Knowing this we are happy using 0.1 mM ABTS because it is saturating slowly and the maximum is not too high. 0.1 mM ABTS is therefore established!<br><br> | ||
| + | |||
| + | ===Wednesday May 30th=== | ||
| + | |||
| + | *'''Team Cloning Bacterial Laccases:''' | ||
| + | :* After plasmid isolation we digested our plasmids with ''Not''I to see if the colony PCR was correct and our laccases are in the backbone. The agarose gel showed that for all of the different plasmids we had at least one plasmid with two DNA bands on the correct height in agarose gel. | ||
| + | |||
| + | ===Thursday May 31st=== | ||
| + | * '''Team Cloning of Bacterial laccase''': | ||
| + | ** We sent the isolated pSB1C3 plasmids with xccl(T7)_His, Bpul(T7)_HIS and Ecol(T7)_His for sequencing. | ||
| + | |||
| + | ===Friday June 1st=== | ||
| + | * '''Team Activity Test''': | ||
| + | ** Today we prepared for our next measurements by setting up the sodium acetate buffer in different pHs. We choose to test the activity of TVEL0 in a pH-Gradient of 1,2,5,7 and 9. | ||
| + | |||
| + | ===Saturday June 2nd=== | ||
| + | ===Sunday June 3rd=== | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** PCRs of genomic DNA on bhal from ''B. halodurans C-125'' we ordered from [http://www.dsmz.de/ DSMZ] before. We handled the cells in the same way we did with ''T. thermophilus'' before and soluted the lyophlized cells in water and boiled them before PCR. After PCR we cleaned up the product with gel electrophoresis and [[Team:Bielefeld-Germany/Protocols/Materials#Used_Kits | PCR clean-up kit]]. However the DNA amount was so low that we had to do the PCRs again. | ||
| + | |||
<html> | <html> | ||
</p> | </p> | ||
| Line 346: | Line 508: | ||
| - | <div><h3>Summary of Week | + | <div><h3>Summary of Week 6</h3> |
<p class="more"> | <p class="more"> | ||
</html> | </html> | ||
| Line 354: | Line 516: | ||
<p> | <p> | ||
</html> | </html> | ||
| - | + | ||
| + | ==Week 6 (06/04 - 06/10/12)== | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
| + | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | ||
| + | <ul> | ||
| + | <li class="toclevel-1 tocsection-1"><a href="#Week_6_.2806.2F04_-_06.2F10.2F12.29"><span class="tocnumber">1</span> <span class="toctext">Week 6 (06/04 - 06/10/12)</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-2 tocsection-2"><a href="#Monday_June_4th"><span class="tocnumber">1.1</span> <span class="toctext">Monday June 4th</span></a></li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-3"><a href="#Tuesday_June_5th"><span class="tocnumber">1.2</span> <span class="toctext">Tuesday June 5th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-4"><a href="#Wednesday_June_6th"><span class="tocnumber">1.3</span> <span class="toctext">Wednesday June 6th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-5"><a href="#Thursday_June_7th"><span class="tocnumber">1.4</span> <span class="toctext">Thursday June 7th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-6"><a href="#Friday_June_8th"><span class="tocnumber">1.5</span> <span class="toctext">Friday June 8th</span></a></li> | ||
| + | <li class="toclevel-2 tocsection-7"><a href="#Saturday_June_9th"><span class="tocnumber">1.6</span> <span class="toctext">Saturday June 9th</span></a></li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-8"><a href="#Sunday_June_10th"><span class="tocnumber">1.7</span> <span class="toctext">Sunday June 10th</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </td></tr></table> | ||
| + | |||
| + | ===Monday June 4th=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | :* The Bacteria ''S. griseus'' and ''S. lavendulae'' has been delivered so we can start with PCRs. We set the first PCR with them as followed: | ||
| + | :** '''PCR table''' | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Material !! Volume | ||
| + | |- | ||
| + | | Buffer (10x Phusion) || 10µL | ||
| + | |- | ||
| + | | Phusion Polymerase || 0,5µL | ||
| + | |- | ||
| + | | dNTPs || 1µL | ||
| + | |- | ||
| + | | Primer Mix || 1µL | ||
| + | |- | ||
| + | | Template DNA || 1µL | ||
| + | |- | ||
| + | | DMSO || 1,5µL | ||
| + | |- | ||
| + | | Water || 35µL | ||
| + | |- | ||
| + | |} | ||
| + | ** ''' PCR program''' | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Temperature !! Time | ||
| + | |- | ||
| + | | 1) 98°C || 7 mins | ||
| + | |- | ||
| + | | 2) 98°C || 20 sec | ||
| + | |- | ||
| + | | 3) 55°C || 20 sec | ||
| + | |- | ||
| + | | 4) 72°C || 1 min | ||
| + | |- | ||
| + | | 5) 72°C || 3 min | ||
| + | |- | ||
| + | | 6) 12°C || | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Cycle between step 2 and 4 35 times. | ||
| + | |||
| + | *'''Team Fungal and Plant Laccases: ''' | ||
| + | **Primerdesign for isolating a laccase from ''Arababidopsis thaliana'' cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag. | ||
| + | |||
| + | ===Tuesday June 5th=== | ||
| + | * '''Team Student Academy:''' | ||
| + | ** Transformation of a plasmid mixture of either pMTE cp46 His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] or pMTE cp46 His and [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] has a more intense fluorescence and was chosen for the experiment. | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence. | ||
| + | ** Again we did PCRs on ''T. thermophilus'' laccase and ''B. halodurans'' laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction. | ||
| + | |||
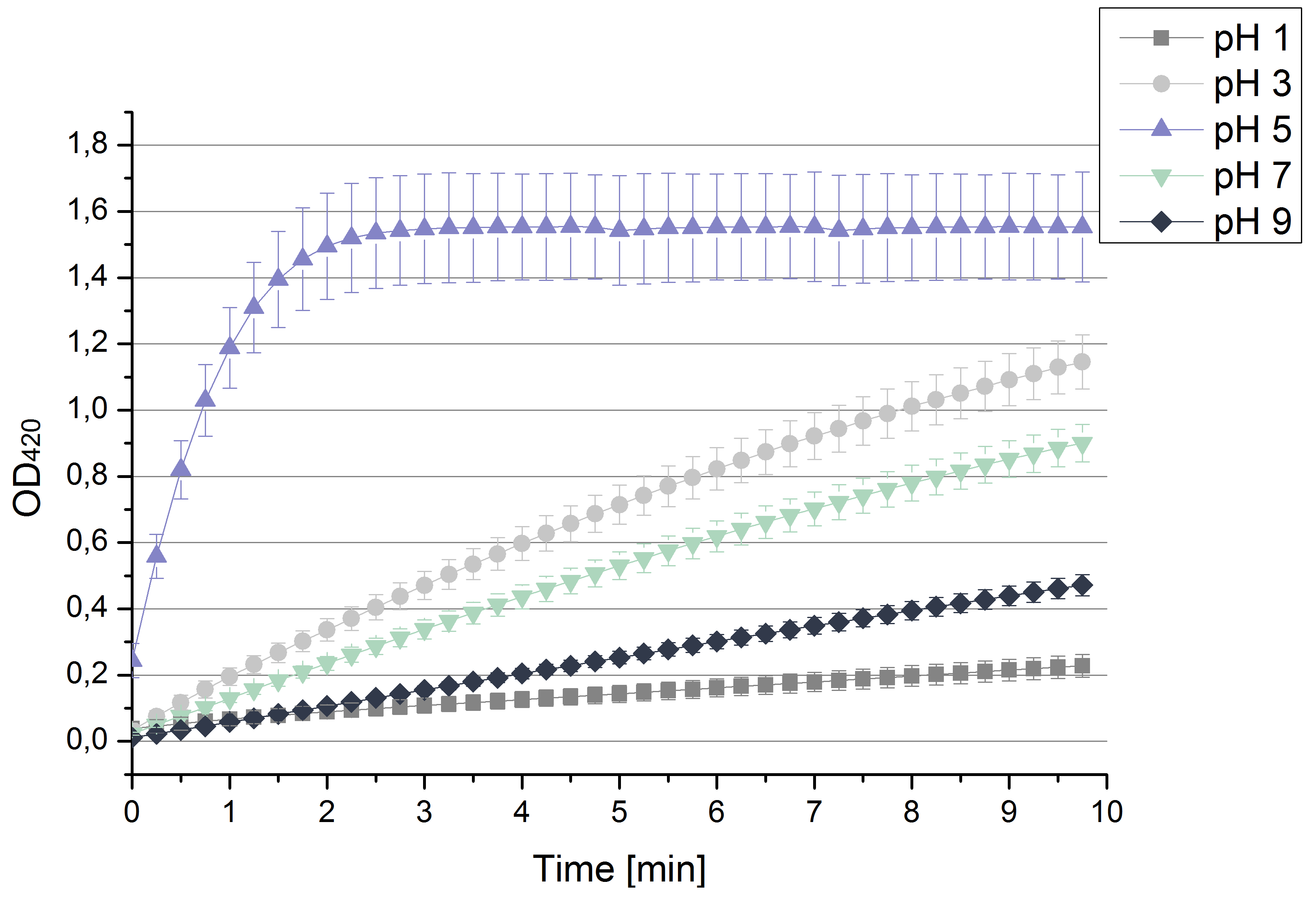
| + | [[File:Bielefeld2012 pHstrametes.png|thumb|right|Activity measurements of the bought laccase from ''T.versiolor'' analyzed through the OD<sub>420</sub> of oxidized ABTS in sodium acetate buffer at different pHs depending on time. Values are calculated by taking the average and standard deviation out of 4 measurements (n=4).]] | ||
| + | * '''Team Activity Tests''': | ||
| + | ** After testing the ''T. versicolor'' laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned. | ||
| + | |||
| + | ===Wednesday June 6th=== | ||
| + | * '''Team Wiki:''' Yay for Team Wiki´s first entry. Our first steps with the iGEM Bielefeld 2012 Wiki contain thinking about contents, layouts, programming and responsibilities. Our first rules are: | ||
| + | ** we are programming static pages in HTML and all the other pages (those that will be updated by all team members) in wiki code. | ||
| + | ** we created all pages and will fill them up with some nice and beautiful content constantly from now on. | ||
| + | ** Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon. | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases:''' Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent ''E. coli'' KRX cells. | ||
| + | |||
| + | ===Thursday June 7th=== | ||
| + | * '''Team Student Academy:''' | ||
| + | ** Repeating of Transformation of 06/05 to verify the function. It is reproducible :) | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands. | ||
| + | |||
| + | * '''Team Modeling''': becoming acquainted with matlab while reading the manual | ||
| + | |||
| + | ===Friday June 8th=== | ||
| + | |||
| + | *'''Team Cloning of Bacterial Laccases:''' | ||
| + | **We plated colonies for plasmid isolations on new plates and made a control restriction with ''Not''I. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His. | ||
| + | |||
| + | ===Saturday June 9th=== | ||
| + | ===Sunday June 10th=== | ||
| + | |||
<html> | <html> | ||
</p> | </p> | ||
| Line 360: | Line 627: | ||
| - | <div><h3>Summary of Week | + | <div><h3>Summary of Week 7</h3> |
<p class="more"> | <p class="more"> | ||
</html> | </html> | ||
| Line 368: | Line 635: | ||
<p> | <p> | ||
</html> | </html> | ||
| - | + | ==Week 7 (06/11 - 06/17/12)== | |
| + | |||
| + | __NOTOC__ | ||
| + | |||
| + | |||
| + | |||
| + | ===Monday June 11th=== | ||
| + | * '''Team Cloning of Bacterial Laccases:''' | ||
| + | **Prepared plasmids for sequencing. We sent another isolated plasmid with'ecol(T7)_His. Also tthl(t7)_His, bahl(T7)_His and bpul(T7)_His plasmids were ready for sequencing. | ||
| + | |||
| + | ===Tuesday June 12th=== | ||
| + | *'''Team Student Academy:''' | ||
| + | ** The whole experiment was tested by another team member to plan the course. | ||
| + | |||
| + | ===Wednesday June 13th=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': Since the GC amount of the ''S. griseus'' and ''S. lavendulae'' laccases are high we used betain to solve the PCR problem. Addition of betain did not change anything on the results, we still didn't got our laccase DNA. | ||
| + | |||
| + | * '''Team Modeling''': Programming our first differential equation and finding the ODE15s function witch solves these equations. | ||
| + | |||
| + | ===Thursday June 14th=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** Because our PCRs have not worked well we thought it may depends on the primer annealing temperature so we did gradient PCR with the same conditions as before ([https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week6 PCR June 4th]). But this also showed no result. Because we made Coloyn PCRs from the arrived DSMZ reaction tubes our next idea was to cultivate the bacteria in media and isolate genomic DNA. | ||
| + | |||
| + | * '''Team Activity Tests''': | ||
| + | ** Since our Tecan microplate reader is not able to actively cool down to 4 °C we got the chance to meet the photometer Carry. Check "protocols" for further information about her. We used the same set up with 100 mM natrium acetate buffer, 0,1 U ''T. versicolor'' laccase and 0,1 mM ABTS as before but now measured at 4°C. Our team is planning to visit a municipal sewage plant for getting some insights into the water conditions there, so we will for sure test other temperatures after having more information. Let´s hope the water there is a little warmer since laccase does not seem to be totally satisfied at 4°C. I would not either. | ||
| + | |||
| + | ===Friday June 15th=== | ||
| + | |||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** Sequencing of the pSB1C3 plasmid with bhal(T7)_His was ok. In conflict to our reference sequence there was a point mutation in the DNA sequence but this mutation doesn’t lead to another amino acid. So..next BioBrick (<partinfo>BBa_K863020</partinfo>) is ready to use! | ||
| + | ** The sequenced plasmid bpul(T7)_His showed again the same mutation in the laccase ORF compared to the reference sequence. We concluded that probably the PCR amplification caused the point mutation. So we did the digest of bpul(T7)_His PCR products from a new PCR, ligated it in pSB1C3 backbone and transformed it in competent KRX cells. Additionally we did the digest tthl(T7)_His and the ligation in pSB1C3 backbone again. | ||
| + | |||
| + | ===Saturday June 16th=== | ||
| + | |||
| + | ===Sunday June 17th=== | ||
<html> | <html> | ||
</p> | </p> | ||
Revision as of 15:52, 23 September 2012
Prologue
Prologue
Starting the team
Beginning in january and february members of the former iGEM team from Bielefeld started seminars to inform interested students about synthetic biology, iGEM and the past Bielefeld projects. In March the final 2012 iGEM Bielefeld team was formed of 15 students and weekly meetings began. Our team was established and it was time to find a suitable project.
Find a project
The first weekly meeting were more like big group brainstorming and we discussed idea, which in some cases were totally different from each other. Everyone had to inform about ideas of others so that, in the end, we all could discuss together.
First project ideas were:
- the detection of multiresistent pathogens
- communication between bacteria and fungi using quorum sensing
- a bacterial hand warmer
- a possibility to detect and destroy mold fungus
- something about spontaneous combustion of hay bale
- an enzyme dispenser
For most of the ideas little information was available. For example spontaneous combustion of hay bales is probably a combination of the metabolisms of different microorganisms and fungus. After some reports in media and press about the environmental effects of steroid hormones, we decided to go for hormones. From the beginning our aim was not to detect but to degrade hormones. We found several possible ways for degradation as there are the hydrolysis of estradiol-derivates with sufatases and glucoronidases. But we thought the best way to degrade steroid hormones would be with the use of laccases. Laccases have the ability to radicalize aromatic rings and can therefore be used to degrade or polymerize a broad range of substances, such as steroid hormones, special insecticides, polycyclic aromatic carbohydrates and aromatic acids. In nature laccases are often used for degradation or polymerisation of lignin or pigments.
Molding together to a team
After we found our project idea we decided to have a get-to-know-weekend with some presentations about iGEM, important methods and ideas for human practices. We also held presentations about other possible iGEM projects to extend our horizon, as there were: e.g. RNA aptamers and magnetotactic bacteria. But the most important part of this weekend was the growing as a team. We realized that we all had one summer to work together, have fun together and most important to stand up together as a team.
Find the right
Now it was time to organize the work and find a suitable task for everyone. In a developing team a lot of different jobs have to be done, e.g.:
- finding sponsors
- communication with the public
- human practices
- wiki- and homepage-design
- modelling
- a forum for exchange of information
- a joker, who entertains the team and lifts the mood
And finally lab work began, feel free to follow us in our weekly labjournal and have a look how our labwork, results and of course problems and their solutions, evolved.
Summary of Week 1
We began our time in the lab with the cultivation of Xanthomonas campestris B100 and E. coli BL21(DE3) to isolate the genomic DNA to do PCRs and purify the desired laccase ORFs. In Order to do this we at first had to designed the PCR-primers. We decided, that the forward primers had to included the prefix, the T7 promotor, the RBS and the first 20 bases of the gene of interest and the reverse primers should consist of the last 20 bases of the gene of interest, a His-tag, followed by two stop codons and the suffix. Furthermore we started making preparations for the Student Academy. The Students Academy is a week-lasting summer school (9th to 13th of July) we got the chance to take part in organizing it. It is distinguished with a lot of presentations and lessons for pupils, but also guided experiments they had to do by themselves. Therefore the general idea for our experimental part was to give the students an understanding of the principle methods in biotechnology and synthetic biology by using fluorescent proteins. So the first step was to searched the Parts Registry for two plasmids with different fluorescent proteins and antibiotic resistances, respectively.
Week 1 (04/30 - 05/06/12)
Contents |
weekly seminar:
- Do we want to order strains of Trametes versicolor and Trametes villosa?
- Gathering information about signal sequences in yeast
- Decision to create a database, so that we can easily number and inscribe our lab results
- Decision to arrange a summer school for pupils in their last year before the final exams
- Discussion about how to meet a member of the german Bundestag (the german parliament)
Monday April 30th
- Team Student Academy: We got the chance to organize one part of the first school academy “synthetic biology/ biotechnology” at the CeBiTec of University Bielefeld by arranging experiments for the pupils and by presenting us and the iGEM competition. For the experimental part our general idea was to give them an understanding of principle methods in biotechnology / synthetic biology by using fluorescent proteins. We planned the following experiments:
- Plasmid isolation of RFP/GFP from a liquid culture.
- Transformation of a plasmid mixture consisting of two different fluorescent proteins (e.g. RFP and GFP) and different antibiotic resistances into E.coli KRX. It will be plated out on LB agar plates without antibiotics and on plates containing one of the two antibiotics, which are present on the plasmids. This way we can demonstrate the effect of antibiotics as selective pressure.
- Team Bacterial Laccases:
- Before our lab time started we sent requests for different plasmids to working groups, which have already worked with laccases we are interested in. Sadly just one working group responded to us. We got answer for a vector with the laccase-ORF CotA from Bacillus pumilus ATCC7061 and a ampicillin resistance from the Swiss Federal Laboratories for Materials Science and Technology, Laboratory for Biomaterials in Switzerland. They promised to send us the plasmid pBpL6. More information...
- In a paper we found a research group who worked with the laccase CopA from Xanthomonas pv. campestris ATCC33913. Luckily the sequence of this laccase is the same in Xanthomonas campestris pv. campestris B100 which we got from a working group at our university. The same thing with a laccase from E. coli. We found papers which described the laccase CueO from E. coli W3110. After blasting this laccase we found out that E. coli BL21(DE3) has this laccase, too. We decided to isolate the laccase from E. coli BL21(DE3).
- Generating new competent E.coli KRX cells.
- Cultivation of Xanthomonas campestris B100 and E. coli BL21(DE3). The bacterial strains we got from a working group at our University. After cultivation we isolated the genomic DNA. The DNA was needed as template for PCRs to purify the wanted laccase ORFs.
- Primer design for isolation of laccases from genomic DNA of Xanthomonas campestris B100 and E. coli BL21(DE3) and for isolation of CotA from Bacillus pumilus ATCC7061 from plasmid. The forward primers were designed with T7 promoter, RBS and the first 20 bases of the wanted gene after prefix. The reverse primers were designed with the last 20 bases of the wanted gene without the stop codon, a HIS-Tag, two stop codons and suffix sequence. Primers: Xcc_LAC_FW_T7, Xcc_LAC_RV_HIS, E.coli_LAC_FW_T7, E.coli_LAC_RV_HIS, B.pumi_LAC_FW_T7 and B. pumi_LAC_RV_HIS
Tuesday May 1th
- Team Student Academy: Searching for two plasmids with different fluorescent proteins behind and antibiotic resistance in parts registry. Found BBa_J04450, a Plasmid with RFP and chloramphenicol resistance (but lacI and CAP sensitive), BBa_J23100, a plasmid with RFP and ampicillin resistance and BBa_I13522, a Plasmid with GFP and ampicillin resistance in Kit Plate 2011.
Wednesday May 2th
- Team Activity Test: Good morning everybody and welcome to the labjournal of Team Activity Tests. Today we started our work with some literature research about enzyme activity tests, laccases and its substrates. So today was filled with online research, reading papers and collecting information about the laccases our team decided to use.
Thursday May 3th
- Team Bacterial Laccases:
- After the vector with the laccase gene CotA from Bacillus pumilus arrived, we transformed it into the competent E.coli KRX which we have already made competent to have a greater amount of vector. The protocol we used was as followed:
- The electroporation setup: U= 2,5kV C= 25 µF and R= 400 <math>\omega</math>
- Since we did not know the efficient of our competent KRX we used two different E.coli volumes for the transformation, 50µL and 100µL. We gave 50µL 10% glycerol to the reaction tubes with 1µL of the vector DNA (Bacillus pumilus). After the transformation we plated them on ampicillin-selection-agar-plates.
- PCR with the Xanthomonas campestris B100 and E. coli BL21(DE3) genomic DNA to isolate the laccases. Therefore we used the primers Xcc_LAC_FW_T7, Xcc_LAC_RV_HIS, E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS which are listed under Materials.
- PCR table
- After the vector with the laccase gene CotA from Bacillus pumilus arrived, we transformed it into the competent E.coli KRX which we have already made competent to have a greater amount of vector. The protocol we used was as followed:
| Material | Volume |
|---|---|
| Buffer (10x Phusion) | 10µL |
| Phusion Polymerase | 0,5µL |
| dNTPs | 1µL |
| Primer Mix | 1µL |
| Template DNA | 1µL |
| DMSO | 1,5µL |
| Water | 35µL |
- PCR program
| Temperature | Time |
|---|---|
| 1) 98°C | 30 sec |
| 2) 98°C | 15 sec |
| 3) 62°C | 45 sec |
| 4) 72°C | 1 min |
| 5) 72°C | 3 min |
| 6) 12°C |
Cycle between step 2 and 4 35 times.
Friday May 4th
Team Bacterial Laccases: We did Colony PCR on the transformed the Bacillus pumilus CotA plasmid. Unfortunately the control with colony PCR didn't work. So we just picked some colonies for plasmid isolation in the hope that on the AMP plate were no false positives colonies.
Summary of Week 2
hier eine Zusammenfassung
Week 2 (05/07 - 05/13/12)
Contents |
weekly seminar:
- Found our first sponsors: Evonik, BioCircle and Merck, now treaties have to be created and signed
- Julia is working on the database
- Decision to organize waver sell to fill up our petty cash
- Gabi and Isabel are designing a poster for the waver sell
- For our human practices we wanted to find a sociology student, willing to think about bioethics, but failed
- Our video is nearly done, is cutted and only needs be underlain with music
Monday May 7th
- Team Student Academy:
- First transformation of BBa_J04450 and BBa_I13522 and plating on selective agar. Result: We got little colonies. There weren’t any green colonies and only some pale red fluorescent colonies.
- Team Cloning of Bacterial Laccases:
- More PCRs of laccase genes xccl from Xanthomonas campestris pv. campestris B100 and ecol from E. coli BL21(DE3) with the isolated genomic DNA as template and Xcc_LAC_FW_T7 / Xcc_LAC_RV_HIS and E.coli_LAC_FW_T7 / E.coli_LAC_RV_HIS primer pairs.
- Since we wanted to screen and characterize laccases from different bacteria we had to order the bacterial strains which weren't available at Bielefeld University from DSMZ. Below is a list of the ordered strains and the laccases we want to isolate from these strains.
- Laccase from Thermus thermophilus HB27 (look here for a publication to this laccase)
- BH2082 from Bacillus halodurans C-125 (look here for a publication to this laccase)
- We ordered S. lavendulae sp. lavendulae ATCC 14158. Originally we wanted the strain Streptomyces lavendulae REN-7 but this strain isn't available at DSMZ. So we now hope that the laccase gene STSL from Streptomyces lavendulae REN-7 is similar to that from S. lavendulae sp. lavendulae ATCC 14158 because there's no DNA sequence for the laccase from this strain available. publication
- We wanted the laccase EpoA from Streptomyces griseus IFO 13350 (for the publication look here. This strain was not available so we ordered Streptomyces griseus ATCC 10137. Unfortunately for this strain are no blast results after blasting the laccase from Streptomyces griseus IFO 13350 against database. So we decided to make primers for the laccase sequence from Streptomyces griseus IFO 13350 in the hope that the sequences are similar enough to get a PCR product.
- Team Modeling:
- Looking for suitable software and enzymkinetics to model the degradation of our substrates with the different laccases. Finding the Michaelis-Menten kinetics and matlab.
Tuesday May 8th
- Team Student Academy:
- Repetition of the transformation didn’t change the result. We made a liquid culture of BBa_J04450, but it did not fluoresce. Searching for mistakes and alternatives. Maybe competent cells are not that good and in case of RFP the lacI sensitivity could be the problem.
- Team Cloning of Bacterial Laccases:
- After some empty agarose gels we finally isolated the laccase gene bpul from Bacillus pumilus ATCC7061 as PCR product with the desired overhanging ends. As template we used the plasmid we got from the Swiss working group.
Wednesday May 9th
- Team Activity Test: From the information we collected during our literature research we created a protocol for our first experiments. We decided to check the activity via a photometer. The one we may use here at the Cebitec is a Tecan Microplate reader. Check protocols for further information. If oxidized by laccase, ABTS can me measured at 420 nm. Also we found out that sodium acetate buffer (100 mM / pH 5) would give an optimal environment to our enzyme. So let´s have a look at our protocol:
- Initial laccase activity test:
- 100 mM sodium acetate buffer, pH 5.0
- 5 mM ABTS
- 8 U laccase
- ad 200 µL deionized H20
- Also we talked about further characterization after accomplishing the first experiments and confirming that the used concentrations are a good choice. We are planning to buy and characterize the laccase from T.versicolor (TVEL0), to have a comparison to our future recombinant laccases. That laccase we are going to analyze in sodium acetate buffers that are adjusted to pH 1, 3, 5, 7 and 9. Further we are going to analyze the effect of different temperatures on the enzymes activity. For that we will first do some more research on the temperatures of the waste water in clarification plants here in Germany. Also we found out that an addition of copper does enhance the laccases activity, so we are going to do some measurements with copper concentrations from 0.1 mM to 0.5 mM in each sample. This seems like some great experiments for the start, so next we are going to order what we need to do the measurements.
Thursday May 10th
- Team Student Academy
- Testing the competent cells by transformation of pUC19. The transformation did not work that good, so that we produced new ones.
- Team Cloning of Bacterial Laccases
- We got the ordered strains from DSMZ. So we did PCR on Thermus thermophilus genomic DNA. First we dissolved some of the lyophilized powder in water and for opening the cells we boiled them for a few minutes. The primers we used were T.thermo_LAC_FW_T7 and T.thermo_LAC_RV_HIS to get the laccase with the same overhangs described in Monday April 30th. Finally with additional DMSO and GC-buffer we had a product of the GC-rich laccase.
Friday May 11th
- Team Activity Tests: For some pre test and characterization for our future laccase activity standard we ordered laccase from Trametes versicolor. As well we had to order a substrate that the laccase could use to demonstrate its abilities. According to the literature ABTS is a well working substrate to characterize oxidizing enzym activity. So we ordered.
Summary of Week 3
hier eine Zusammenfassung
Week 3 (05/14 - 05/20/12)
Contents |
weekly seminar:
- first lab service: Robert
- our GFP, which we wanted to use for the summer school for pupils, does not work
- first competent cells have to be made: Julia S. and Robert
- Decision to buy a commercial laccase to establish the analytics and the enzyme tests
- our expose has to be translated into english: Malak
- last planning for our waver sell
- Julia S. is creating a vector for Pichia pastoris and is now looking for sequences
- The BMBF invites all german iGEM teams to Berlin to attend at the Biotechnologie2020+ strategy process
Monday May 14th
Tuesday May 15th
Wednesday May 16th
- Team Cloning of Bacterial Laccases:
- For cloning our laccases we need pSB1C3 backbone. Therefore we we transformed BBa_J04450 (pSB1C3 with RFP) in competent KRX cells.
Thursday May 17th
- Team Cloning of Bacterial Laccases: Plasmid isolation of <partinfo>BBa_J04450</partinfo>.
- Team Modeling:
- Meeting Mrs. Lutter, a mathematics prof. of our course of studies and looking for our first model of a metabolic pathway, finding out, that we don't need such a complex model. Start thinking that we want and what we need.
Friday May 18th
- Team Activity Tests: Our T.versicolor laccase and the ABTS arrived! We couldn´t wait to start, so we set up the stock solutions we will need, such as sodium acetat buffer (pH 5), 10 mM ABTS and deluted laccase.
Saturday May 19th
Sunday May 20th
Summary of Week 4
hier eine Zusammenfassung
Week 4 (05/21 - 05/27/12)
Contents |
weekly seminar:
- Lab service: Isabel
- We try to establish a collaboration with the iGEM team from SDU-Denmark
- Got our distribution kits
- First successful cloning and cultivations
- Who wants to be a summer school teacher?
- We will not travel to the ACHEMA because only local teams are invited
- Do we want to participate in the Biolympics? (It's a sports party with fun organized by the bts)
Monday May 21st
- Team Cloning of Bacterial Laccases:
- We wanted to clone our laccase PCR products xccl and ecol in pSB1C3 backbone. Therefore we did some restriction digests on the PCR products and the vector BBa_J04450.
- Team Modeling: Our aims for modeling:
- model the expression of the laccases in the organisms.
- model the activity of our enzymes.
- model the interesting parts of a clarification plant (the part witch are interesting for our cleaner.
Tuesday May 22nd
- Team Cloning of Bacterial Laccases:
- Ligation of the digested PCR products in pSB1C3 backbone and transformation in KRX electro-competent cells.
Wednesday May 23rd
- Team Student Academy:
- Repetition of the transformation of BBa_J04450 and BBa_I13522 with new competent E. coli KRX cells. Got intense red fluorescing colonies but no green fluorescing colonies. Made a backup of E. coli KRX BBa_J04450.
- Asking for other plasmids containing GFP at the working groups of our University.
- Team Cloning of Bacterial Laccases:
- After there were no colonies on our pSB1C3 + xccl(T7)_His (Xanthomonas campestris) transformation plate we did the transformation with the same ligation preparation again. The other ligation with pSB1C3 + ecol(T7)_His (E. coli) showed colonies so we started colony PCRs to find positive colonies. Sadly the colony PCRs showed no products but the problem was that we just had the long overhang primers (E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS ). Therefore we ordered the F and R primers.
Thursday May 24th
- Team Student Academy:
- Made a liquid culture of E. coli KRX with BBa_J04450 at 30 °C. There was no fluorescence.
- Transformation of BBa_J23100 into E. coli KRX. Also got intense red fluorescing colonies.
- Team Activity Tests:
- On our schedule today was testing our bought laccases and improving our protocol in case it won't work. We got familiar with the flat bottom microplates and got a briefing into using the Tecan correctly. We started with the following setup: 100 mM sodium acetate (pH 5), 5 mM ABTS, 8 U TVEL0 laccase, ad 200 µL with deionized water. Then we measured the absorption at 420 nm every 15 seconds over a time period of 2 minutes. Immediately our samples turned dark blue but unfortunately the changeover was out of range to be detected by Tecan. With this information we needed to reduce the laccase concentration to get measurable results. We have chosen to try another measurement with 0.1 U TVEL0 laccase and it worked! The result was a nice saturation curve but it reached a too high, because not good measurable, optimum within 1 minute (Fig. 1). To avoid the loss of important data in the beginning of the reaction and to reduce the saturation OD we decided to slow everything down by using less ABTS.
Friday May 25th
- Team Cloning of Bacterial Laccases:
- We had to do the PCR on T. thermo laccase again because after the purification of the last PCR product the DNA amount was very low.
- Team Student Academy:
- Made a liquid culture of E. coli KRX with BBa_J23100 at 30 °C. Result: There was no fluorescence.
Saturday May 26th
Sunday May 27th
- Team Activity Test:
- Remember our first activity measurements? The activity was awesome and reached quickly its saturation. So today we decided to have a lazy Sunday and reduce absorption maximum and reaction speed, too. For this we used 0.1 mM ABTS instead of 5 mM ABTS and we got good results. The saturation reached its maximum at a OD420 of ~1.6 after roundabout 2 minutes (see Fig. 2). Taking together we halved the maximal OD420 and doubled the reaction time until maximum is reached with using 0.1 mM ABTS instead of 5 mM ABTS.
Summary of Week 5
hier eine Zusammenfassung
Week 5 (05/28 - 06/03/12)
Contents
|
Monday May 28th
- Team Student Academy:
- Made liquid cultures of E. coli KRX with BBa_J04450 and with BBa_J23100. Result: Intense red fluorescence. We made a glycerol stock and a plasmid isolation
- Team Cloning of Bacterial Laccases:
- Digest of ecol(T7)_HIS, xccl(T7)_His and bpul(T7)_His and BBa_J04450 (pSB1C3 + RFP) with the same restriction enzymes, ligation of the digested products and transformation in E. coli KRX cells.
- Since we have less bpul(T7)_His and tthl(T7)_His DNA, we set an PCR from the remaining PCR approach.
Tuesday May 29th
- Team Student Academy:
- E. coli Mach1 with pMTE cp46 His received from the working group “Fermentation Engineering”, University Bielefeld. Plasmid contains genes for GFP and kanamycine resistance. We plated it and made a liquid culture at 37°C. Result: There was an intense fluorescence. We made a glycerol stock and a plasmid isolation.
- Team Cloning of Bacterial Laccases:
- We did colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the E. coli laccase, the Xanthomonas campestris laccase and the B. pumilus laccase with the inducible T7 promoter and a His-tag.
- Team Activity Tests:
- After establishing our recipe for activity measurements we were curious about different ABTS concentration and wanted to make sure we took the right one for our approach. With this in mind we did our activity measurement with 0.1 U TVEL0 laccase, 100 mM sodium acetate buffer (pH 5), ad 200 µl H2O dest., but with two new ABTS concentrations, namely 0.2 mM and 0.05 mM ABTS (see Fig. 1). It turned out that using 0.05 mM ABTS leads to a low maximum of saturation but reaches it quickly. With 0.2 mM ABTS the opposite occurs: the activity curve is saturated at a OD420 of ~2.7 but it needs more time to reach its maximum. Knowing this we are happy using 0.1 mM ABTS because it is saturating slowly and the maximum is not too high. 0.1 mM ABTS is therefore established!
- After establishing our recipe for activity measurements we were curious about different ABTS concentration and wanted to make sure we took the right one for our approach. With this in mind we did our activity measurement with 0.1 U TVEL0 laccase, 100 mM sodium acetate buffer (pH 5), ad 200 µl H2O dest., but with two new ABTS concentrations, namely 0.2 mM and 0.05 mM ABTS (see Fig. 1). It turned out that using 0.05 mM ABTS leads to a low maximum of saturation but reaches it quickly. With 0.2 mM ABTS the opposite occurs: the activity curve is saturated at a OD420 of ~2.7 but it needs more time to reach its maximum. Knowing this we are happy using 0.1 mM ABTS because it is saturating slowly and the maximum is not too high. 0.1 mM ABTS is therefore established!
Wednesday May 30th
- Team Cloning Bacterial Laccases:
- After plasmid isolation we digested our plasmids with NotI to see if the colony PCR was correct and our laccases are in the backbone. The agarose gel showed that for all of the different plasmids we had at least one plasmid with two DNA bands on the correct height in agarose gel.
Thursday May 31st
- Team Cloning of Bacterial laccase:
- We sent the isolated pSB1C3 plasmids with xccl(T7)_His, Bpul(T7)_HIS and Ecol(T7)_His for sequencing.
Friday June 1st
- Team Activity Test:
- Today we prepared for our next measurements by setting up the sodium acetate buffer in different pHs. We choose to test the activity of TVEL0 in a pH-Gradient of 1,2,5,7 and 9.
Saturday June 2nd
Sunday June 3rd
- Team Cloning of Bacterial Laccases:
- PCRs of genomic DNA on bhal from B. halodurans C-125 we ordered from DSMZ before. We handled the cells in the same way we did with T. thermophilus before and soluted the lyophlized cells in water and boiled them before PCR. After PCR we cleaned up the product with gel electrophoresis and PCR clean-up kit. However the DNA amount was so low that we had to do the PCRs again.
Summary of Week 6
hier eine Zusammenfassung
Week 6 (06/04 - 06/10/12)
Contents
|
Monday June 4th
- Team Cloning of Bacterial Laccases:
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
- PCR table
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
| Material | Volume |
|---|---|
| Buffer (10x Phusion) | 10µL |
| Phusion Polymerase | 0,5µL |
| dNTPs | 1µL |
| Primer Mix | 1µL |
| Template DNA | 1µL |
| DMSO | 1,5µL |
| Water | 35µL |
- PCR program
| Temperature | Time |
|---|---|
| 1) 98°C | 7 mins |
| 2) 98°C | 20 sec |
| 3) 55°C | 20 sec |
| 4) 72°C | 1 min |
| 5) 72°C | 3 min |
| 6) 12°C |
Cycle between step 2 and 4 35 times.
- Team Fungal and Plant Laccases:
- Primerdesign for isolating a laccase from Arababidopsis thaliana cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag.
Tuesday June 5th
- Team Student Academy:
- Transformation of a plasmid mixture of either pMTE cp46 His and BBa_J04450 or pMTE cp46 His and BBa_J23100. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. BBa_J23100 has a more intense fluorescence and was chosen for the experiment.
- Team Cloning of Bacterial Laccases:
- The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence.
- Again we did PCRs on T. thermophilus laccase and B. halodurans laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction.
- Team Activity Tests:
- After testing the T. versicolor laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned.
Wednesday June 6th
- Team Wiki: Yay for Team Wiki´s first entry. Our first steps with the iGEM Bielefeld 2012 Wiki contain thinking about contents, layouts, programming and responsibilities. Our first rules are:
- we are programming static pages in HTML and all the other pages (those that will be updated by all team members) in wiki code.
- we created all pages and will fill them up with some nice and beautiful content constantly from now on.
- Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon.
- Team Cloning of Bacterial Laccases: Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent E. coli KRX cells.
Thursday June 7th
- Team Student Academy:
- Repeating of Transformation of 06/05 to verify the function. It is reproducible :)
- Team Cloning of Bacterial Laccases: Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands.
- Team Modeling: becoming acquainted with matlab while reading the manual
Friday June 8th
- Team Cloning of Bacterial Laccases:
- We plated colonies for plasmid isolations on new plates and made a control restriction with NotI. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His.
Saturday June 9th
Sunday June 10th
Summary of Week 7
hier eine Zusammenfassung
Week 7 (06/11 - 06/17/12)
Monday June 11th
- Team Cloning of Bacterial Laccases:
- Prepared plasmids for sequencing. We sent another isolated plasmid with'ecol(T7)_His. Also tthl(t7)_His, bahl(T7)_His and bpul(T7)_His plasmids were ready for sequencing.
Tuesday June 12th
- Team Student Academy:
- The whole experiment was tested by another team member to plan the course.
Wednesday June 13th
- Team Cloning of Bacterial Laccases: Since the GC amount of the S. griseus and S. lavendulae laccases are high we used betain to solve the PCR problem. Addition of betain did not change anything on the results, we still didn't got our laccase DNA.
- Team Modeling: Programming our first differential equation and finding the ODE15s function witch solves these equations.
Thursday June 14th
- Team Cloning of Bacterial Laccases:
- Because our PCRs have not worked well we thought it may depends on the primer annealing temperature so we did gradient PCR with the same conditions as before (PCR June 4th). But this also showed no result. Because we made Coloyn PCRs from the arrived DSMZ reaction tubes our next idea was to cultivate the bacteria in media and isolate genomic DNA.
- Team Activity Tests:
- Since our Tecan microplate reader is not able to actively cool down to 4 °C we got the chance to meet the photometer Carry. Check "protocols" for further information about her. We used the same set up with 100 mM natrium acetate buffer, 0,1 U T. versicolor laccase and 0,1 mM ABTS as before but now measured at 4°C. Our team is planning to visit a municipal sewage plant for getting some insights into the water conditions there, so we will for sure test other temperatures after having more information. Let´s hope the water there is a little warmer since laccase does not seem to be totally satisfied at 4°C. I would not either.
Friday June 15th
- Team Cloning of Bacterial Laccases:
- Sequencing of the pSB1C3 plasmid with bhal(T7)_His was ok. In conflict to our reference sequence there was a point mutation in the DNA sequence but this mutation doesn’t lead to another amino acid. So..next BioBrick (<partinfo>BBa_K863020</partinfo>) is ready to use!
- The sequenced plasmid bpul(T7)_His showed again the same mutation in the laccase ORF compared to the reference sequence. We concluded that probably the PCR amplification caused the point mutation. So we did the digest of bpul(T7)_His PCR products from a new PCR, ligated it in pSB1C3 backbone and transformed it in competent KRX cells. Additionally we did the digest tthl(T7)_His and the ligation in pSB1C3 backbone again.
Saturday June 16th
Sunday June 17th
Summary of Week 3
hier eine Zusammenfassung
hier text
Summary of Week 3
hier eine Zusammenfassung
hier text
Summary of Week 3
hier eine Zusammenfassung
hier text
Summary of Week 3
hier eine Zusammenfassung
hier text
Summary of Week 3
hier eine Zusammenfassung
hier text
| 55px | | | | | | | | | | |
 "
"