Team:Amsterdam/summary
From 2012.igem.org
Summary
The ever increasing complexity of genetic circuits will soon call for the need of enhanced biological data storage devices. In light of this we have devised an innovative generic memory system that couples to the signals in these circuits. This Cellular Logbook can remember which of these signals were encountered at what intensity or at which time. Storage is done by DNA methylation of restriction sites, after which we can read out whether or not a signal has been present by use of restriction enzymes and gel electrophoresis.
Furthermore, because of the dilution of methylated bits due to cellular proliferation and degradation, an indication of the time of registering can be inferred from the proportian of methylated bits in the total bits. This novel memory system not only offers great expandability in terms of detectable signals, but also holds great promise as a multi-sensor utility in experimental and industrial environments.
Experimental Setup
A preliminary methylation-based plasmid, lacking the Zinc Finger, was constructed using the Gibson assembly method. A lambda pLac hybrid promoter (R0011), medium ribosomal binding site (B0032), M.ScaI methyltransferase attached to a Myc linker and a double terminator (B0014) were cloned into the pSB1AT3 backbone plasmid (origin of replication pUC19-derived pMB1, copy number 100–300 per cells, ampicillin and tetracycline resistance genes). The final construct containing two ScaI sites was transformed into Library Efficiency DH5α™ Competent bacteria (Invitrogen Cat. No. 18263-012) and were cultured overnight in LB medium and ampicillin, in the presence and absence of 100 µM IPTG. The plasmid was isolated and digested with the restriction enzymes EcoRI, ScaI and ScaI + EcoRI (double digestion), incubated for an hour at 37˚C and run on a 1% agarose gel in 0.5X TBE with 0.5 µgml-1 ethidium bromide.
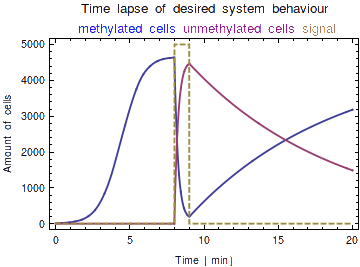
Model: Memory Generations
Methylation patterns are not transferred to the progeny in bacterial cell division, which will cause the amount of methylated bits after signal registering to be diluted over time due to natural cell death and plasmid degradation. Because of this, the time of onset of the signal can be inferred from the fraction of methylated bits / total bits. A ‘toy model’ established the desired working of our system and yielded insight into the roles of various parameters therein (e.g. proliferation rate, death rate, processing speed). Toy model: 1.A single bit per cell, which can either be in either the methylated or unmethylated state 2.Logistic (bounded) growth for the memory cell population, with a capacity limit of 5000. 3.A fast processing speed, defined as the rate at which bits are written in response to signal detection, causing all cells to be methylated within a short time window. 4.Division of a methylated cell results in a methylated and an unmethylated cell
Caption: By measuring the fraction of methylated/total bits, the time of offset of the signal can be deduced. Analyzing the above graph, we see a unique fraction for each time point after the onset of the signal.
 "
"