Team:Amsterdam/project/growthrates
From 2012.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
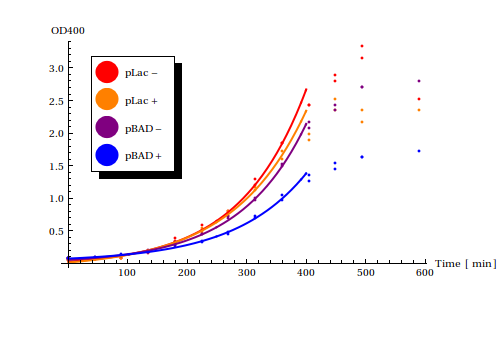
Both experiments were performed in cell strains <math>\text{DH5}\alpha</math>. | Both experiments were performed in cell strains <math>\text{DH5}\alpha</math>. | ||
| - | [[File:rateestimation.png|frame|Growth rates of two different constructs (pLac, pBAD) with either the corresponding signal (lactose, arabninose) present or not present]] | + | [[File:rateestimation.png|frame|400px|Growth rates of two different constructs (pLac, pBAD) with either the corresponding signal (lactose, arabninose) present or not present]] |
Using the Mathematica function <code>NonLinearModelFit</code> functions of the form <math>a + b e^{c t}</math>, with <math>t</math> as time in minutes, were fitted to the exponential phases of the growth curves. | Using the Mathematica function <code>NonLinearModelFit</code> functions of the form <math>a + b e^{c t}</math>, with <math>t</math> as time in minutes, were fitted to the exponential phases of the growth curves. | ||
| Line 31: | Line 31: | ||
Suprisingly, pBAD reduces the growth rate more strongly than pLac. | Suprisingly, pBAD reduces the growth rate more strongly than pLac. | ||
pLac is known to be a better stronger promoter than pBAD with higher leaky expression rates and was thus expected to have a stronger negative effect on the growth rate. | pLac is known to be a better stronger promoter than pBAD with higher leaky expression rates and was thus expected to have a stronger negative effect on the growth rate. | ||
| - | |||
This results suits us very well! As you can read <here> pBAD was also shown to have lower leaky expression rates in our own experiments and thus functions more robustly. | This results suits us very well! As you can read <here> pBAD was also shown to have lower leaky expression rates in our own experiments and thus functions more robustly. | ||
Revision as of 09:46, 23 September 2012
 "
"