Team:Amsterdam/data/experimental
From 2012.igem.org
(Difference between revisions)
| (4 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<div id="content-area"> | <div id="content-area"> | ||
<div id="sub-menu" class="content-block"> | <div id="sub-menu" class="content-block"> | ||
| - | <h1>Experimental | + | <h1>Experimental Results</h1> |
| - | + | __NOTOC__ | |
<div class='clear'></div> | <div class='clear'></div> | ||
<h2>Functionality of the writer-reader module under different sensor modules</h2> | <h2>Functionality of the writer-reader module under different sensor modules</h2> | ||
| Line 16: | Line 16: | ||
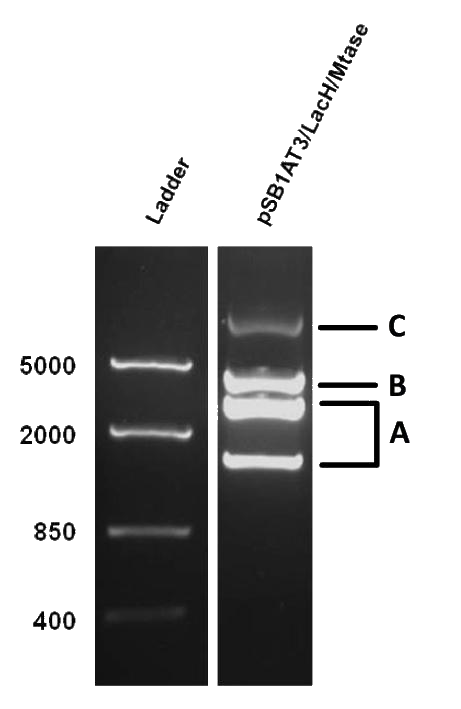
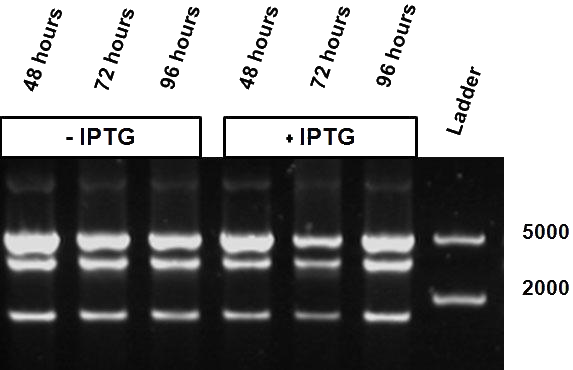
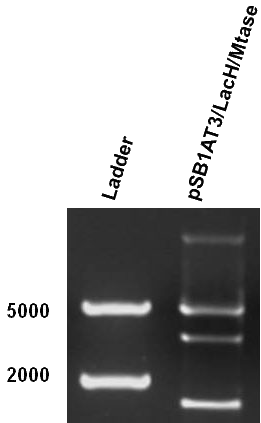
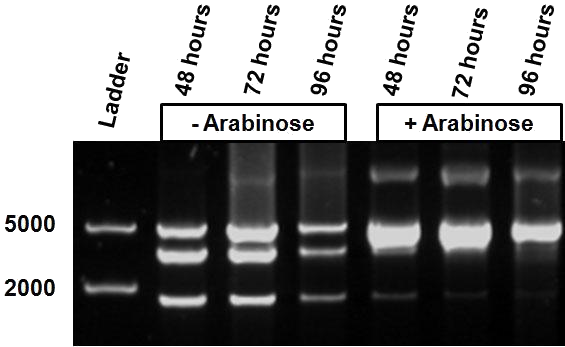
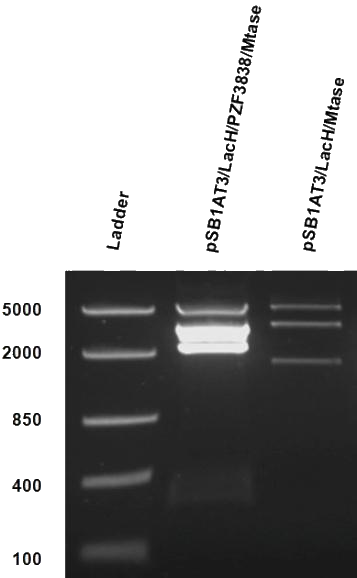
As mentioned in Molecular design, the ScaI restriction enzyme is unable to cut methylated restriction sites. Therefore, we expect different possible restriction profiles through ScaI restriction digestion since there is one ScaI site residing in the pSB1AT3 backbone and we created one ScaI site via a scar inside our writer module. We expect to find either an off or intermediate methylation state knowing that the LacH promoter driving the MTase has some basal activity. | As mentioned in Molecular design, the ScaI restriction enzyme is unable to cut methylated restriction sites. Therefore, we expect different possible restriction profiles through ScaI restriction digestion since there is one ScaI site residing in the pSB1AT3 backbone and we created one ScaI site via a scar inside our writer module. We expect to find either an off or intermediate methylation state knowing that the LacH promoter driving the MTase has some basal activity. | ||
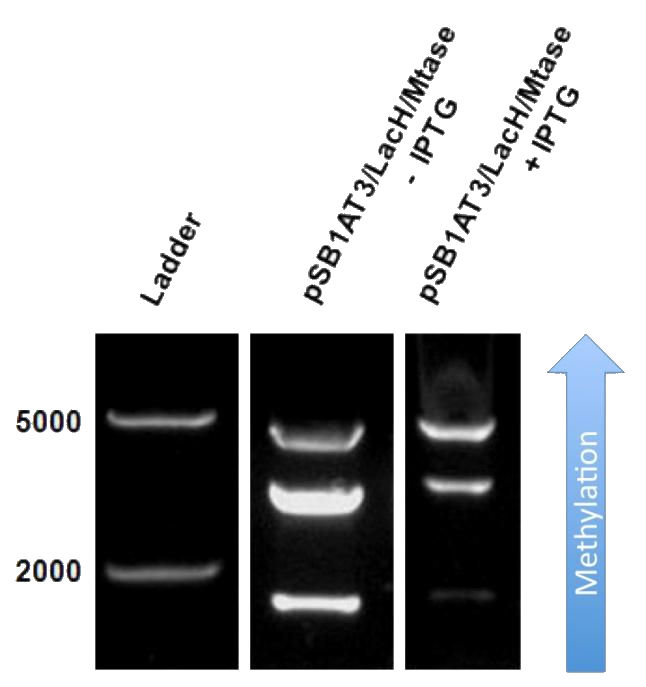
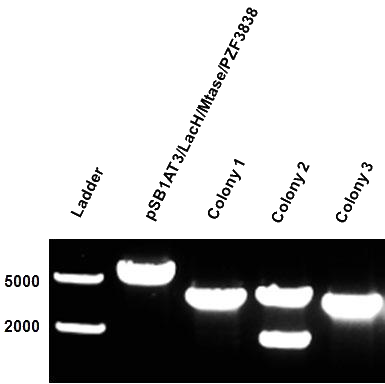
| - | Figure 1 shows the result of a ScaI restriction digestion of the pSB1AT3/LacH/MTase construct without IPTG induction. The pattern displayed corresponds to a combination of bands indicating an intermediate state between the ‘off’ and ‘on’ situation. Interpretation of the read-out: absence of MTase expression in E. | + | Figure 1 shows the result of a ScaI restriction digestion of the pSB1AT3/LacH/MTase construct without IPTG induction. The pattern displayed corresponds to a combination of bands indicating an intermediate state between the ‘off’ and ‘on’ situation. Interpretation of the read-out: absence of MTase expression in ''E. coli'' leads to a complete digestion of our plasmid. Two bands of 2989 bp and 1621 bp are then observed (A). Incomplete methylation of the plasmid at only one of the two ScaI sites shows a linearized plasmid band 4610 bp. Complete methylation of our writer module prevents ScaI to cut and a typical uncut plasmid profile is observed (C). |
<h4>Behavior of the writer-reader module under IPTG induction</h4> | <h4>Behavior of the writer-reader module under IPTG induction</h4> | ||
| Line 77: | Line 77: | ||
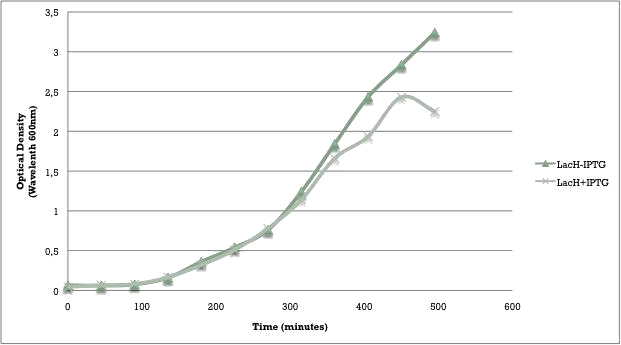
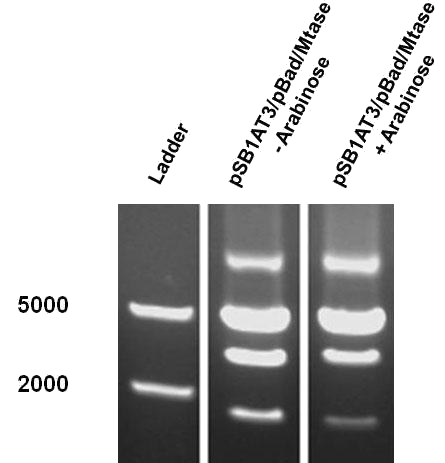
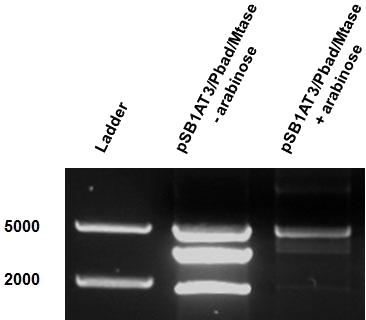
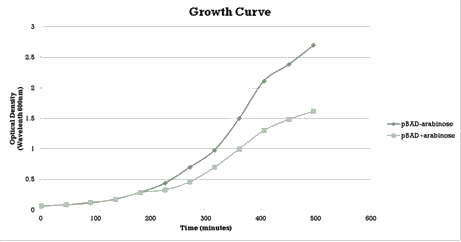
The “off” state was not achieved in the absence of arabinose. All the experiments have been conducted into the high copy number plasmid pSB1AT3. It is known that leaky expression is relative to the copy number of the plasmid.1 Hence, the inability of the Cellular Logbook to show an “off” state in the absence of arabinose could be attributed to the high copy number plasmid used in these experiments as was discussed with the LacH. | The “off” state was not achieved in the absence of arabinose. All the experiments have been conducted into the high copy number plasmid pSB1AT3. It is known that leaky expression is relative to the copy number of the plasmid.1 Hence, the inability of the Cellular Logbook to show an “off” state in the absence of arabinose could be attributed to the high copy number plasmid used in these experiments as was discussed with the LacH. | ||
| - | Similarly to the pSB1AT3/LacH/Mtase, a growth curve experiment was conducted to characterize the acquired construct of pSB1AT3/pBad/Mtase. | + | Similarly to the pSB1AT3/LacH/Mtase, a growth curve experiment was conducted to characterize the acquired construct of pSB1AT3/pBad/Mtase (figure 10). |
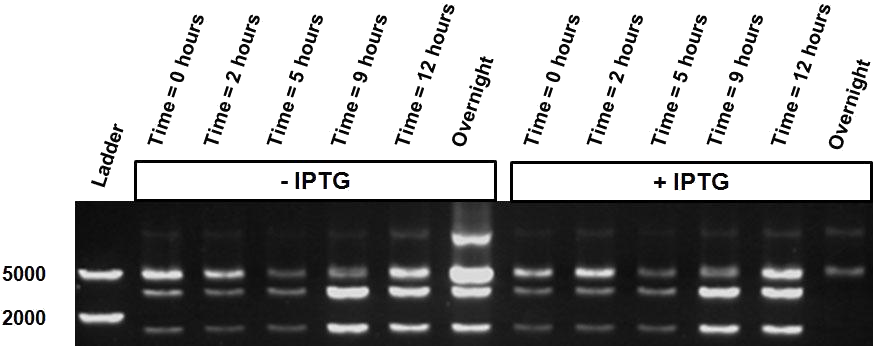
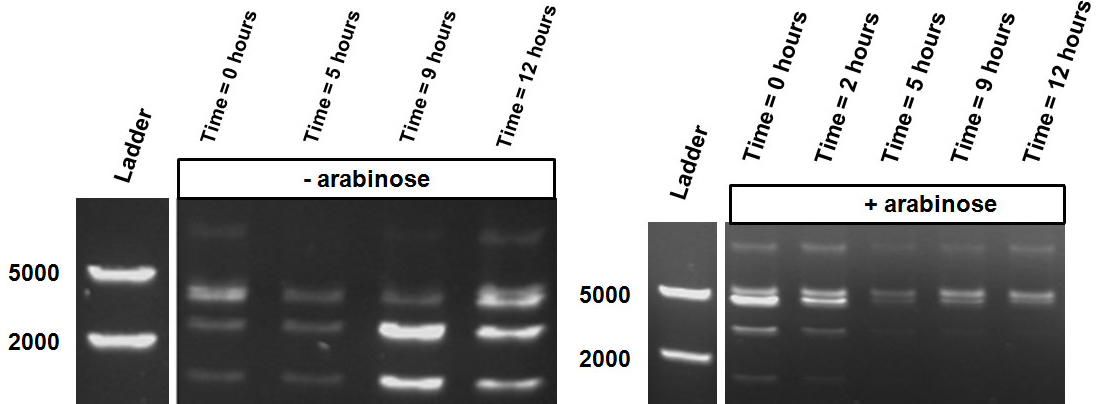
| - | Over the course of time the <br\> methylation-dependent restriction profile observed in the gel showed a shift towards the ‘on’ digestion profile in the presence of arabinose. This result shows that our Cellular Logbook is able to sense and write an arabinose signal present in the medium in a shorter period of time.<br\><br\><br\><br\><br\> | + | Over the course of time the <br\> methylation-dependent restriction profile observed in the gel showed a shift towards the ‘on’ digestion profile in the presence of arabinose (figure 11). This result shows that our Cellular Logbook is able to sense and write an arabinose signal present in the medium in a shorter period of time.<br\><br\><br\><br\><br\> |
<h2>Towards the Cellular Logbook</h2> | <h2>Towards the Cellular Logbook</h2> | ||
Latest revision as of 03:52, 27 September 2012
 "
"