Team:Amsterdam/data/background activity
From 2012.igem.org
(Difference between revisions)
| (8 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div id="sub-menu" class="content-block"> | <div id="sub-menu" class="content-block"> | ||
| - | + | __NOTOC__ | |
| + | <h1>Background Activity</h1> | ||
Here we will propose a solution to the the background noise observed in the experiments, and conclude what rates of leakiness are acceptable to the system. | Here we will propose a solution to the the background noise observed in the experiments, and conclude what rates of leakiness are acceptable to the system. | ||
In this, we model the system both using both ordinary differential equations and stochastically using the Gillespie SSA algorithm. | In this, we model the system both using both ordinary differential equations and stochastically using the Gillespie SSA algorithm. | ||
We will focus on the effects of two rate constants in particular on the fraction of methylated plasmids in absence of the inducing signal: | We will focus on the effects of two rate constants in particular on the fraction of methylated plasmids in absence of the inducing signal: | ||
| - | * the leaky transcription rate of the mRNA for the | + | * the leaky transcription rate of the mRNA for the writer, denoted by $k_{c\text{MW}}$ and |
| - | * the catalysis rate of the | + | * the catalysis rate of the writer, the rate with which it methylates umethylated plasmids |
| - | + | ||
== Introduction == | == Introduction == | ||
| - | Before starting the experiments, a lot of crucial parameters on the molecular system were still unknown. Both the leaky expression rate of the | + | Before starting the experiments, a lot of crucial parameters on the molecular system were still unknown. |
| - | * A basal methylation activity that was always recorded even in absence of the signal.This background noise reduces the dynamic range of our system; there is no way to tell whether observed band intensities are due to the signal having passed the ''Cellular Logbook'' [[Team:Amsterdam/modelling/timeinferrance|some time ago]] or simply because of the background noise. We conclude that storing a signal using the ''Cellular Logbook'' requires a transcriptional regulator that has a high repressing quality in order to diminish the background activity. | + | Both the leaky expression rate of the sensor and the catalysis rate of the writer were hard to model because of this. |
| + | Sensible predictions on how the molecular system we were designing was going to function exactly therefore seemed impossible. | ||
| + | We did expect the promoter/sensor we were going to test, pLac, to show some leaky expression. As to how much background activity this was going to cause, we couldn't predict and thus we proceeded to test out the construct in the lab. The [[Team:Amsterdam/Lab|laboratory results]] seemed to suffer from two problems: | ||
| + | * A basal methylation activity that was always recorded even in absence of the signal. This background noise reduces the dynamic range of our system; there is no way to tell whether observed band intensities are due to the signal having passed the ''Cellular Logbook'' [[Team:Amsterdam/modelling/timeinferrance|some time ago]] or simply because of the background noise. We conclude that storing a signal using the ''Cellular Logbook'' requires a transcriptional regulator that has a high repressing quality in order to diminish the background activity. | ||
* A lack of a fully induced state for the pLac operon, but not for the later tested pBAD operon. We suspect this problem to be caused by a low uptake rate of the signalling compound (lac operon inducer IPTG) compared to the amount of repressor LacI present in the system controlling the operons. Especially for the part characterization experiments in which the pLac-MTase construct was cloned into a high copy number plasmid, this could be a problem. This problem is solved by adding the ''LacY''-gene that codes for permease to the existing construct. Inclusion of this gene might also lead to higher background methylation percentages in the negative controls, however. | * A lack of a fully induced state for the pLac operon, but not for the later tested pBAD operon. We suspect this problem to be caused by a low uptake rate of the signalling compound (lac operon inducer IPTG) compared to the amount of repressor LacI present in the system controlling the operons. Especially for the part characterization experiments in which the pLac-MTase construct was cloned into a high copy number plasmid, this could be a problem. This problem is solved by adding the ''LacY''-gene that codes for permease to the existing construct. Inclusion of this gene might also lead to higher background methylation percentages in the negative controls, however. | ||
==== Stochastic Differential Equations ==== | ==== Stochastic Differential Equations ==== | ||
| - | + | We expected the basal activity of the used sensors to result in small amounts of proteins and thus thought a stochastic modelling approach would be suitable. | |
| - | When trying to accurately model these small amounts of proteins the thermodynamic equilibrium assumption that characterizes ODE-models might not be | + | When trying to accurately model these small amounts of proteins the thermodynamic equilibrium assumption that characterizes macroscopic ODE-models might not be maintainable. |
| - | + | In these cases a finer-grained modelling method is required: the mesoscopic Stochastic Simulation Algorithm by Gillespie. | |
| - | In these cases a finer-grained modelling method is | + | By modelling each individual molecular reaction separately, the discreteness of this small amounts system is accounted for. |
| - | By modelling each individual molecular reaction separately, the discreteness of this small amounts system is accounted for. | + | Despite the small amounts of species present in the system analyzed here, we found no qualitative differences between the ODE and stochastic versions of the model. |
==== Comparison of SSA implementations ==== | ==== Comparison of SSA implementations ==== | ||
| - | + | We've investigated many stochastic simulation packages with implementations of the direct algorithms and some more coarse-grained optimizations thereof (e.g. tau-leaping). | |
| - | Most notable are [www.xlr8r.info/SSA/ xSSA] for Mathematica and [http://www.stompy.sourceforge.net StochPy] for Python. | + | Most notable are [http://www.xlr8r.info/SSA/ xSSA] for Mathematica and [http://www.stompy.sourceforge.net StochPy] for Python. |
| - | The former seems a great tool, but at the time of writing seems to lack the ability to process third order reactions (reactions with three reactants). | + | The former seems to be a great tool, but at the time of writing seems to lack the ability to process third order reactions (reactions with three reactants). |
The latter has been developed at the Free University Amsterdam by Timo Maarleveld, currently PhD-student there. | The latter has been developed at the Free University Amsterdam by Timo Maarleveld, currently PhD-student there. | ||
In using this package, we've kept in close contact with Timo and submitted a lot of bug reports, helping the software to grow. | In using this package, we've kept in close contact with Timo and submitted a lot of bug reports, helping the software to grow. | ||
| - | Additionally, we've also extended StochPy by writing a supplementary tool [Team:Amsterdam/ | + | Additionally, we've also extended StochPy by writing a supplementary tool [https://2012.igem.org/Team:Amsterdam/extra/software MDL2LaTeX], that eases the publishing of models developed with StochPy by converting the model input files to LaTeX representations. |
=Leakiness and background noise= | =Leakiness and background noise= | ||
| + | |||
| + | This model has as its main goal to identify how much the expression rate of the uninduced sensor and the catalysis rate of the writer affect the methylation status of the plasmids. | ||
| + | Each plasmid is assumed to have one bit and one operon that controls the gene for the writer. | ||
| + | Running the stochastic version of this model, we noticed that keeping the amount of equations to a minimum is paramount to success, as the computation time seems to increase exponentially with the amount of reactions in the system. | ||
| + | Focussing on the leakiness rate instead of the repressor-operon interactions that would require a lot of equations, we will therefore make one bold assumption: | ||
| + | all operons present in a single cell are permanently bound to a transcriptional repressor. | ||
| + | This frees us from having to code the signal entering the cell and binding the repressing TF, granting focus to the leaky expression rate and the catalysis rate of the fusion protein. | ||
| + | For these models to have any forecastable value therefore, the amount of repressor molecules present in the system has to be much greater than the amount required to cover all operons. | ||
| + | |||
| + | In order to not exclude the possible qualitative difference in model behaviour when all molecular reactions are computed individually, the system has also been modelled stochastically. To make the two systems perfectly comparable, the used rates are qualitatively identical to the ones used in the ODE system. | ||
| + | Furthermore, the same parameter values have been used. | ||
| + | |||
| + | ==ODE Model definition== | ||
<table align="right"> | <table align="right"> | ||
<tr><th>Parameter</th><th>Value</th></tr> | <tr><th>Parameter</th><th>Value</th></tr> | ||
<tr><td>Ca</td> <td>$200\ \text{or}\ 40$</td> | <tr><td>Ca</td> <td>$200\ \text{or}\ 40$</td> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>ksW</td> <td>$30$</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>lMW</td> <td>$0.462$</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td> | + | <td>lW</td> <td>$0.2$</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 60: | Line 76: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
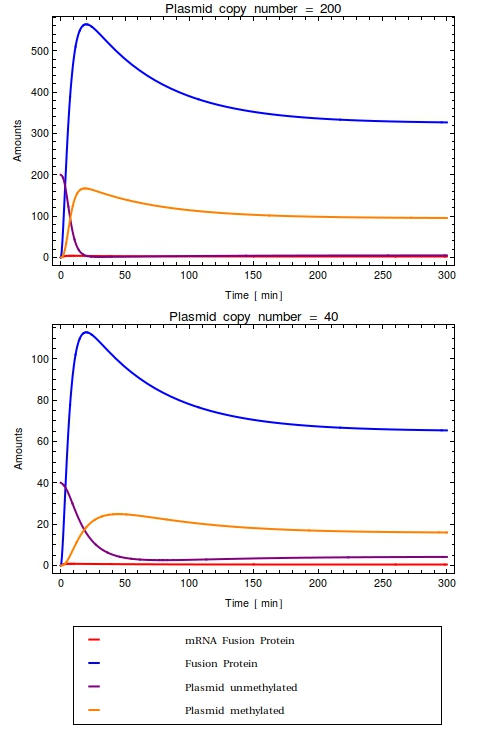
[[File:Combinedode.png|thumb|right|300px|Time trajectories of ODE-model in which the construct has been inserted in a high copy number plasmid (top, $Ca = 200$) and a low copy number plasmid (bottom, $Ca = 40$). All initial species values are set to $0$, except for the intial value of unmethylated plasmids which is equal to $Ca$. With these parameters, the dynamic range of the system is completely taken up by the background noise. No qualitative difference is observed between the high and low copy number cases]] | [[File:Combinedode.png|thumb|right|300px|Time trajectories of ODE-model in which the construct has been inserted in a high copy number plasmid (top, $Ca = 200$) and a low copy number plasmid (bottom, $Ca = 40$). All initial species values are set to $0$, except for the intial value of unmethylated plasmids which is equal to $Ca$. With these parameters, the dynamic range of the system is completely taken up by the background noise. No qualitative difference is observed between the high and low copy number cases]] | ||
| - | + | Writer-mRNA results only from leaky expression from the operons $O$ with leakiness rate $k_{\text{sMW}}$ and is degraded with rate $\lambda_{\text{MW}}$: | |
$$ | $$ | ||
| - | \frac{d\text{ | + | \frac{d\text{mW}}{dt} = k_{\text{sMW}} \cdot \text{O} - \lambda_{\text{MW}} \cdot \text{MW} |
$$ | $$ | ||
| - | + | Writer is created from mRNA and with rate $k_{\text{sW}}$ and degraded with rate $\lambda_{\text{W}}$: | |
$$ | $$ | ||
| - | \frac{d\text{ | + | \frac{d\text{W}}{dt} = k_{\text{sW}} \cdot \text{MW} - \lambda_{\text{W}} \cdot \text{W} |
$$ | $$ | ||
Unmethylated plasmids are created from both unmethylated and methylated plasmids; methylation is not transferred to daughter plasmid during plasmid replication in prokaryotes. Plasmid growth continues until the plasmid copy number $Ca$ has been reached: | Unmethylated plasmids are created from both unmethylated and methylated plasmids; methylation is not transferred to daughter plasmid during plasmid replication in prokaryotes. Plasmid growth continues until the plasmid copy number $Ca$ has been reached: | ||
$$ | $$ | ||
| - | \frac{d\text{PlasU}}{dt} = k_{\text{Plas}} (1 - \frac{(\text{PlasU} + \text{PlasM})}{Ca}) - k_{ | + | \frac{d\text{PlasU}}{dt} = k_{\text{Plas}} (1 - \frac{(\text{PlasU} + \text{PlasM})}{Ca}) - k_{cW} \cdot \text{W} \cdot \text{PlasU} - \lambda_{\text{Plas}} \cdot \text{PlasU}, |
$$ | $$ | ||
Methylated plasmids result from the fusion protein finding an umethylated plasmid and methylating it: | Methylated plasmids result from the fusion protein finding an umethylated plasmid and methylating it: | ||
$$ | $$ | ||
| - | \frac{d\text{PlasM}}{dt} = k_{c\text{ | + | \frac{d\text{PlasM}}{dt} = k_{c\text{W}} \cdot \text{W} \cdot \text{PlasU} - \lambda_{\text{PlasM}} \cdot \text{PlasM} |
$$ | $$ | ||
The amount of (repressed) operons is identical to the amount of plasmids currently in the system: | The amount of (repressed) operons is identical to the amount of plasmids currently in the system: | ||
| Line 97: | Line 99: | ||
\text{O} = \text{PlasU} + \text{PlasM} | \text{O} = \text{PlasU} + \text{PlasM} | ||
$$ | $$ | ||
| + | |||
| + | == Stochastic model definition == | ||
| + | |||
| + | The equations in the stochastic model are qualitatively equal to the ones in the differential equation model. | ||
| + | The equations, parameter and initial species values can be viewed on [[Team:Amsterdam/achievements/stochastic_model_definition|this page]]. | ||
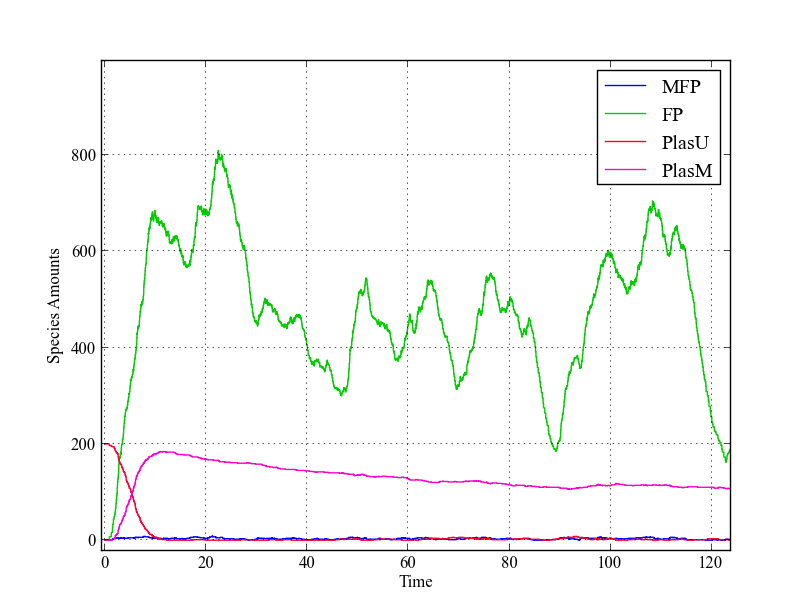
| + | Analyzing the behaviour of this model by looking at the time-lapse plots, we see a similar trend as in the ODE-model: | ||
| + | all plasmids are methylated within a short amount of time. The fusion protein amounts are shown to be widely varying, but this does not alter the fraction of methylated plasmids much. | ||
| + | |||
| + | [[File:Stoch_single.png|thumb|200px|Single trajectory of stochastic model. Just as in the deterministic model, all plasmids are methylated within a short amount of time due to the leaky expression with the used values for $ k_{cat} $ and $ k_{cW} $]] | ||
== Retrieving sensible parameter values == | == Retrieving sensible parameter values == | ||
| Line 113: | Line 124: | ||
We immediately see that the $[O_{\text{Free}}]$ depends on $[R_{2}]$ and $O_{\text{Total}}$. | We immediately see that the $[O_{\text{Free}}]$ depends on $[R_{2}]$ and $O_{\text{Total}}$. | ||
| - | The dissociation constant of the dimerized LacI-repressor for its operator sequence has been determined to be $10^{-12} M $ and the volume of ''E. coli'' is $0.7\ \mu m^{3}$. Assuming $ | + | The dissociation constant of the dimerized LacI-repressor for its operator sequence has been determined to be $10^{-12} M $ and the volume of ''E. coli'' is $0.7\ \mu m^{3}$. Assuming $O_{\text{Total}} = 200$ and $R_{2} = 200$, the time fraction during which $O$ is free is $2.10815 \cdot 10^{-6}$. |
More simply, the leaky expression rate can also be retrieved from this [http://oregonstate.edu/instruction/bb492/lectures/Regulation.html website], which states a 1000-fold decrease in expression of the repressor-bound operon compared to the free operon. | More simply, the leaky expression rate can also be retrieved from this [http://oregonstate.edu/instruction/bb492/lectures/Regulation.html website], which states a 1000-fold decrease in expression of the repressor-bound operon compared to the free operon. | ||
| Line 119: | Line 130: | ||
==== Fusion protein catalysis rate ==== | ==== Fusion protein catalysis rate ==== | ||
The catalysis rate $k_{cat}$ has not been determined for the MTase that we are using, M.ScaI. | The catalysis rate $k_{cat}$ has not been determined for the MTase that we are using, M.ScaI. | ||
| - | However, for another MTase with the same [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.1.1.113 EC2.1.1.113 number] as M.ScaI, BamH1, the $k_{cat}$ has been determined to be $0.0175$ ([[#Cheng| | + | However, for another MTase with the same [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.1.1.113 EC2.1.1.113 number] as M.ScaI, BamH1, the $k_{cat}$ has been determined to be $0.0175$ ([[#Cheng|Cheng (1999)]]). |
==== Cell growth rate ==== | ==== Cell growth rate ==== | ||
| - | Our own experiments showed that the bacterial strain $\text{ | + | Our own experiments showed that the bacterial strain $\text{DH}5\alpha$ transformed with two of our preliminary constructs had growth rates (<math>\mu</math>) between $80\ min^{-1}$ and $90\ min^{-1}$ ([https://2012.igem.org/Team:Amsterdam/project/growthrates growth rates]). |
| - | In these simulations, cellular division was assumed to be either constant at 80 | + | In these simulations, cellular division was assumed to be either constant at 80. |
| - | Looking at the plots for both the high and the low copy number plasmids, no qualitative difference is observable. All species in the system simply reach a steady state defined as | + | Looking at the plots for both the high and the low copy number plasmids, no qualitative difference is observable. All species in the system simply reach a steady state defined as the fraction between their production and degradation rates. |
Most notably, in the steady state that this system reaches all plasmids are methylated. | Most notably, in the steady state that this system reaches all plasmids are methylated. | ||
This is very undesirable of course! | This is very undesirable of course! | ||
== Steady state parameter scanning == | == Steady state parameter scanning == | ||
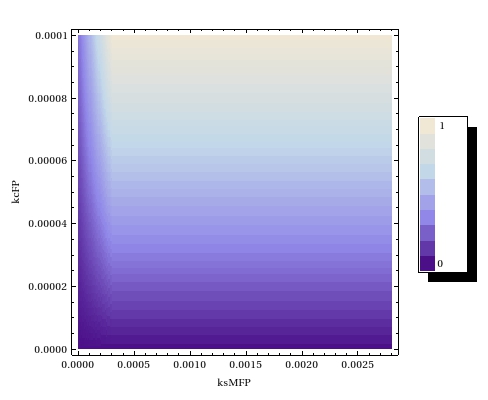
| - | In order to deduce what ranges of operon leaky expression and | + | In order to deduce what ranges of operon leaky expression and W catalysis rates would yield acceptable leaky expression, a steady state parameter scan has been performed. |
| - | [[File:Ssparmscanode.jpeg|thumb|300px|right|Parameter scan to study the effects of the operon leaky expression rate $k_{ | + | [[File:Ssparmscanode.jpeg|thumb|300px|right|Parameter scan to study the effects of the operon leaky expression rate $k_{s0MW}$ and the W catalysis rate $k_{c\text{W}}$ on the steady state fraction of $ \frac{\text{Methylated plasmids}}{\text{Methylated} + \text{Unmethylated plasmids}}$ in the ODE-model]] |
Our experimental results can thus be explained by any combination of the two parameters which which has an intermediate value in plot. | Our experimental results can thus be explained by any combination of the two parameters which which has an intermediate value in plot. | ||
| - | + | A similar parameter scan has been painstakingly performed with the stochastic model. Simulations with 20 trajectories of 200-minute simulations for each parameter combination in a two-dimensional scan for both the $k_{cat}$ and $k_{cW}$ in the same ranges as the ODE parm-scan. Unfortunately, even for these high amount of replications the variability in the resulting methylation fractions is very high, such that no trends are discernible at all in the results (data not shown, python scripts downloadable at the bottom of this page). This variability can probably be reduced by performing even longer simulations and using more trajectories. A much more efficient implementation of the SSA will have to be used to reach this goal within reasonable time scales however. We hope to finish these simulations before the Jamboree. Because the deterministic simulations show similar qualitative behaviour as the more realistic stochastic simulations in the time-lapse plots, one could argue that we will obtain similar results here as the much less computationally expensive ODE-parameter scan. | |
| - | The | + | The steady state fraction of methylation shows almost no dependence upon the leaky transcription rate in the ODE model. This is likely caused by the steady state value that the writer mRNA reaches fairly quickly. The parameter scan density plots shows a very weak to no influence of the background noise. The fact that the degradation rate of the mRNA is much larger than the leaky transcription rate probably causes this. On the contrary, the steady state fraction of methylated plasmids is shown to be heavily dependent upon the catalysis rate of the fusion protein. This can steer future experiments to research the effects of lowered catalysis rates. |
| - | + | = Discussion = | |
| - | + | Unlike our initial expectations, the ODE-model analyzed here suggests that the catalysis rate of the W is more important to the high background noise we have observed in the experiments than the leaky expression rate. | |
| - | + | This yields a new hypothesis to test in the lab: will lowering the catalysis rate significantly lower the background methylation rate? | |
| - | + | A few methods to do this come to mind to try and lower the catalysis rate of the writer protein: | |
| - | + | * Lower the temperature during the experiments, which will slow down all reactions in the cell including the catalysis rate of the writer | |
| - | + | * Mutate amino acid sequence of binding domain, making the affinity of the for the DNA higher | |
| - | + | * Pick a different methyltransferase, one that has a lower catalysis rate | |
| + | * Attach a fluorescent protein to the methyltransferase. This will both increase its diffusion coefficient, decreasing the catalysis rate, and as an added bonus give more insight into the transcription rate of the sensor. | ||
| + | Concluding, we found that the catalysis rate of the writer protein is more important to the high background activity we've observed in the lab. This lead us to suggest several new experiments of which we can hopefully try one or more before the Jamboree! | ||
| - | + | =Source files= | |
| - | + | Source files for both the SSA and ODE model, including the stochastic and ODE parm scanners, are [https://www.dropbox.com/sh/ijn9smzthdf27t5/rkpRFci6qU available over here]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=References= | =References= | ||
| Line 288: | Line 170: | ||
</sup> | </sup> | ||
</span> | </span> | ||
| - | |||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
{{Team:Amsterdam/Foot}} | {{Team:Amsterdam/Foot}} | ||
Latest revision as of 03:53, 27 September 2012
 "
"