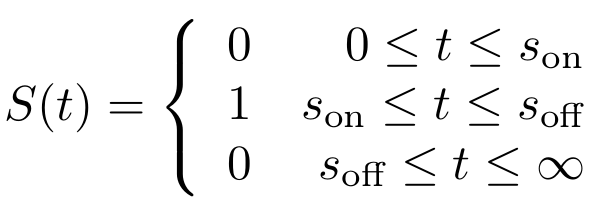Team:Amsterdam/achievements/time inference model
From 2012.igem.org
(Difference between revisions)
(Created page with "{{Team:Amsterdam/stylesheet}} {{Team:Amsterdam/scripts}} {{Team:Amsterdam/Header}} {{Team:Amsterdam/Sidebar1}} <div id="content-area"> <div id="sub-menu" class="content-block"> ...") |
|||
| (2 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div id="sub-menu" class="content-block"> | <div id="sub-menu" class="content-block"> | ||
| + | <h1>Inferring the time of signal onset</h1> | ||
| + | |||
| + | __TOC__ | ||
Using the here presented model, we will examine how to infer the signal detection time from the amount of methylated plasmids. The cellular division rate determines how long a signal is stored in the ''Cellular Logbook''. | Using the here presented model, we will examine how to infer the signal detection time from the amount of methylated plasmids. The cellular division rate determines how long a signal is stored in the ''Cellular Logbook''. | ||
All units are dimensionless in this model, as its sole purpose is clarification of practical usage of the ''Cellular Logbook''. | All units are dimensionless in this model, as its sole purpose is clarification of practical usage of the ''Cellular Logbook''. | ||
| + | <div class='clear'></div> | ||
| - | + | <h2>Methylated bits over time</h2> | |
| - | + | ||
| - | + | ||
Numerous identical plasmids are often present in single cells and plasmids replicate independently of the bacterial chromosome (Scott 1984). A plasmid copy number (PCN) has been determined for all plasmids in the Parts Registry, which indicates a likely amount of copies of the plasmid to be present in each cell. Unlike eukaryotes, prokaryotes do not copy DNA methylation patterns to the newly synthesized strand during DNA replication. This will lead to a dilution of the amount of ‘written’-plasmids over time, mostly due to cell replication and the ensuing binomial division of the plasmids in the parent cell among the two daughter cells. | Numerous identical plasmids are often present in single cells and plasmids replicate independently of the bacterial chromosome (Scott 1984). A plasmid copy number (PCN) has been determined for all plasmids in the Parts Registry, which indicates a likely amount of copies of the plasmid to be present in each cell. Unlike eukaryotes, prokaryotes do not copy DNA methylation patterns to the newly synthesized strand during DNA replication. This will lead to a dilution of the amount of ‘written’-plasmids over time, mostly due to cell replication and the ensuing binomial division of the plasmids in the parent cell among the two daughter cells. | ||
| Line 24: | Line 26: | ||
at the time of memory read-out, the time at which the signal was registered can be inferred. | at the time of memory read-out, the time at which the signal was registered can be inferred. | ||
| - | + | <h2>Model definition</h2> | |
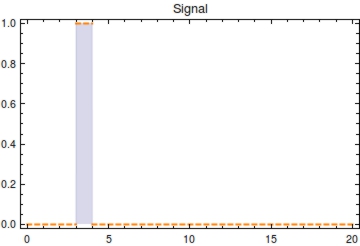
First, let’s model the input signal/compound which is to be reported on. Imagine the to be stationary and positioned along a fluidic stream so that the signal to be registered can pass the ''Cellular Logbook''. Modelling the signal using the piecewise function <math>S(t)</math> now seems appropriate. Here, <math>s_{\text{on}}</math> is defined as the time at which the signal is first encountered and <math>s_{\text{off}}</math> as the time at which the signal is turned off. | First, let’s model the input signal/compound which is to be reported on. Imagine the to be stationary and positioned along a fluidic stream so that the signal to be registered can pass the ''Cellular Logbook''. Modelling the signal using the piecewise function <math>S(t)</math> now seems appropriate. Here, <math>s_{\text{on}}</math> is defined as the time at which the signal is first encountered and <math>s_{\text{off}}</math> as the time at which the signal is turned off. | ||
| Line 109: | Line 111: | ||
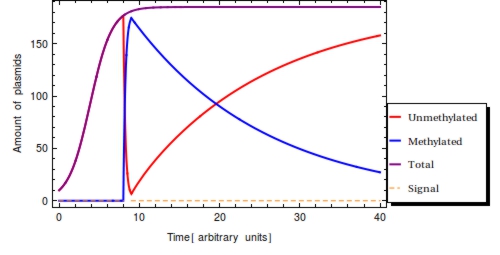
Assuming that the plasmid population will have reached its stationary state level before is plausible and eases the analysis somewhat. If the capacity limit has not been reached yet before, a lower value of results than had the capacity limit been reached. This could fool an experimentalist into thinking that the signal was detected relatively long ago, when in fact the amount of plasmids was still very low at <math>s_{\text{on}}</math>, such that total plasmid population <math>P_{\text{T}}</math> has continued to expand since <math>s_{\text{on}}</math>. | Assuming that the plasmid population will have reached its stationary state level before is plausible and eases the analysis somewhat. If the capacity limit has not been reached yet before, a lower value of results than had the capacity limit been reached. This could fool an experimentalist into thinking that the signal was detected relatively long ago, when in fact the amount of plasmids was still very low at <math>s_{\text{on}}</math>, such that total plasmid population <math>P_{\text{T}}</math> has continued to expand since <math>s_{\text{on}}</math>. | ||
| - | + | <h2>In theory</h2> | |
<center> | <center> | ||
<html> | <html> | ||
| Line 136: | Line 138: | ||
$$ | $$ | ||
| - | + | <h2>In practice</h2> | |
[[File:bands.jpeg|center|thumb|500px|Gel representations for a range of different <math>F(t)</math> values. Complete methylation of all bits results in a single, bright band at the top of the gel. This indicates the undigested, linearized plasmid. Decreasing the amount of methylated bits shifts the intensity of the top band away to the two bottom bands. These indicate the linearized & successfully digested plasmid]] | [[File:bands.jpeg|center|thumb|500px|Gel representations for a range of different <math>F(t)</math> values. Complete methylation of all bits results in a single, bright band at the top of the gel. This indicates the undigested, linearized plasmid. Decreasing the amount of methylated bits shifts the intensity of the top band away to the two bottom bands. These indicate the linearized & successfully digested plasmid]] | ||
Latest revision as of 17:51, 26 September 2012
 "
"