Team:Amsterdam/achievements/experimental
From 2012.igem.org
(Difference between revisions)
| Line 26: | Line 26: | ||
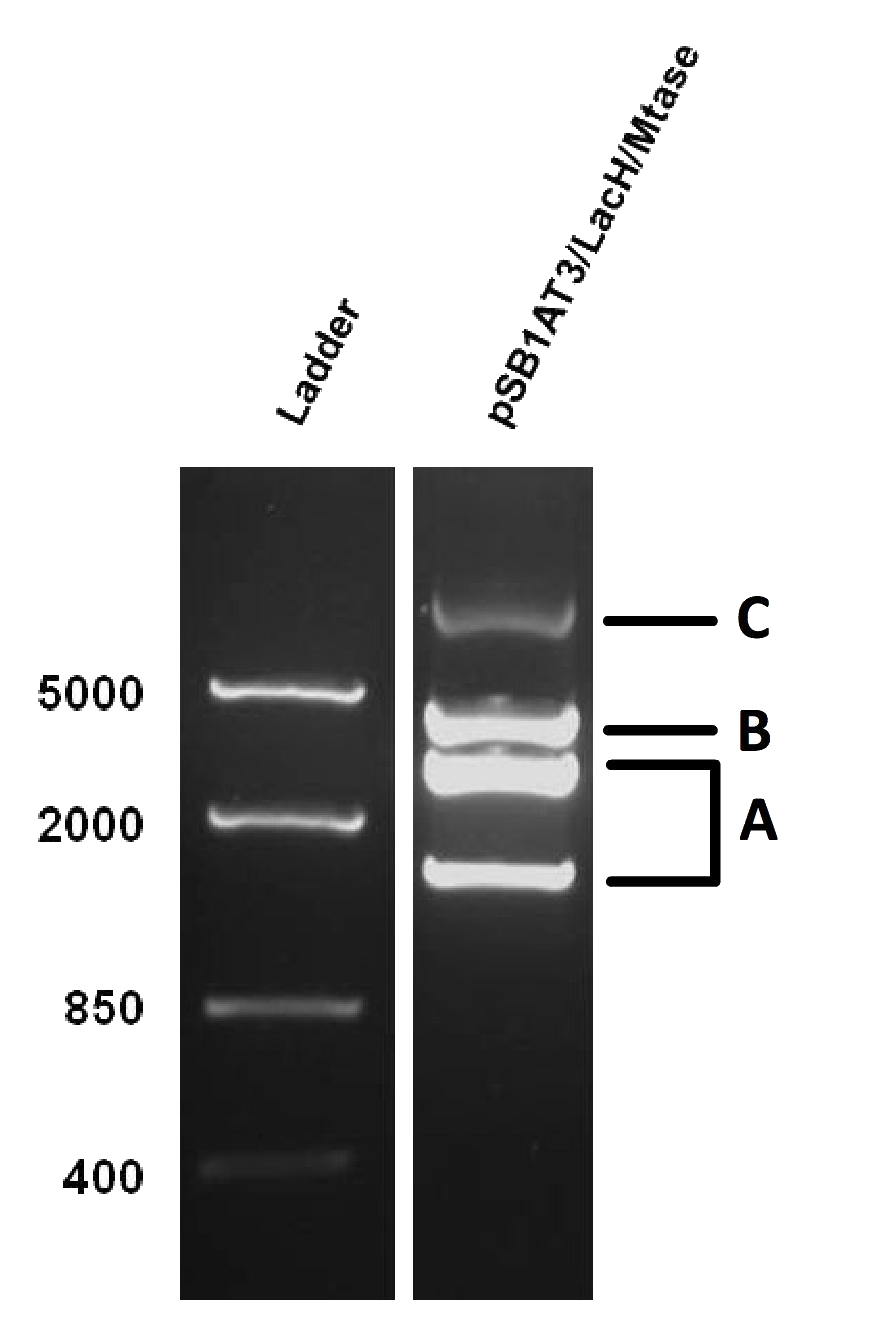
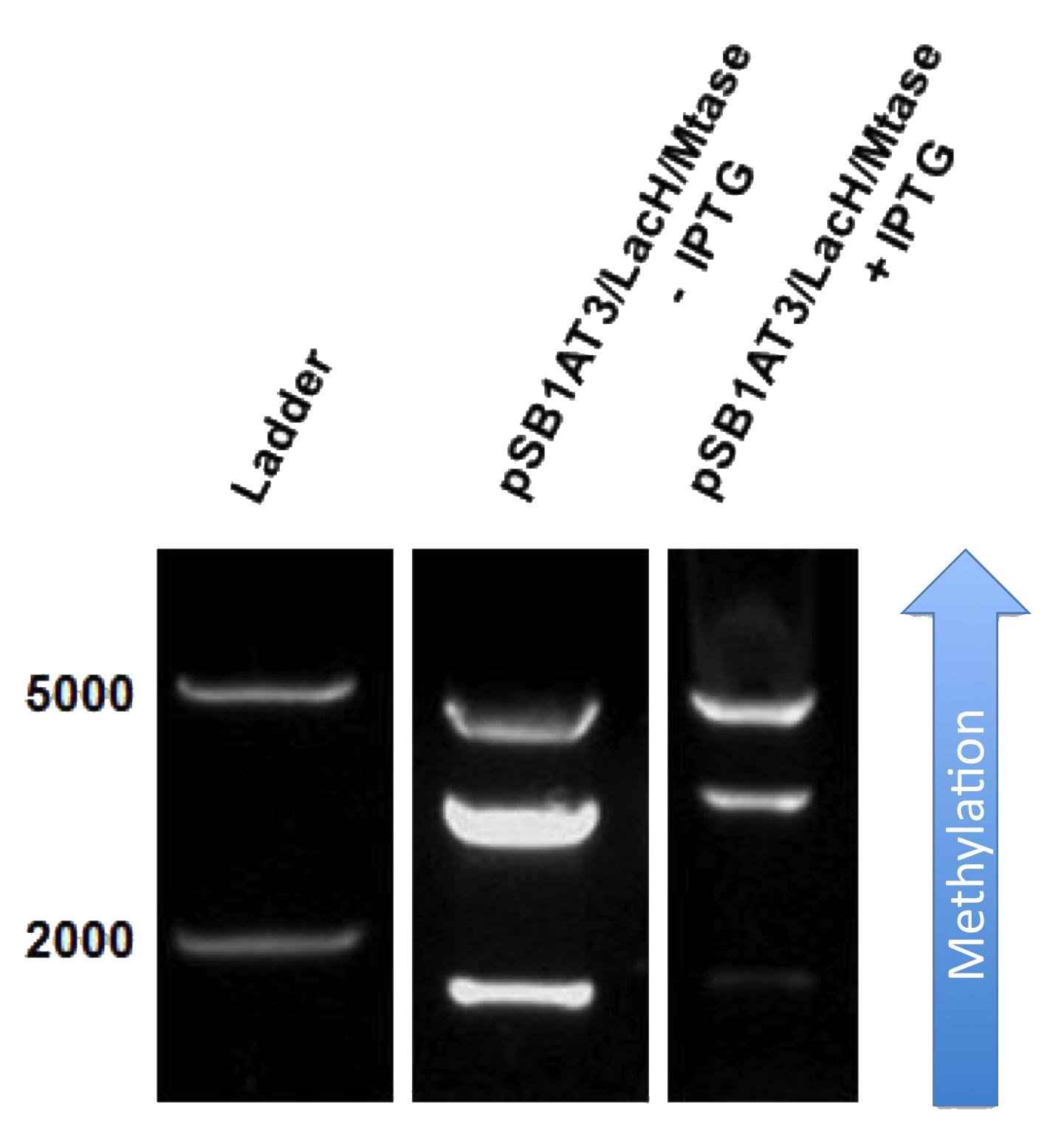
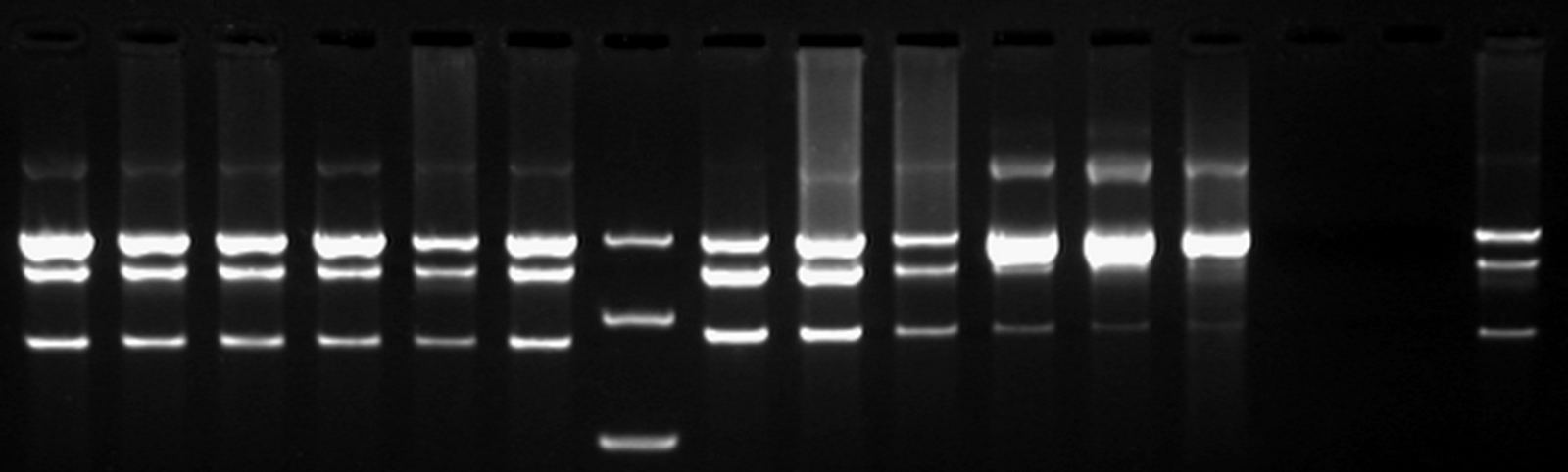
The next step was to see if IPTG-based induction of MTase expression would modify the methylation state of our biobrick thus changing the resulting restriction profile. In the presence of 1mM IPTG, the promoter should be activated providing MTase production inside the cells. Since our [model] for this experiment suggests that methylation occurs at a fast rate we expect that after 16h of IPTG induction, the read-out will dramatically shift to the uncut profile. | The next step was to see if IPTG-based induction of MTase expression would modify the methylation state of our biobrick thus changing the resulting restriction profile. In the presence of 1mM IPTG, the promoter should be activated providing MTase production inside the cells. Since our [model] for this experiment suggests that methylation occurs at a fast rate we expect that after 16h of IPTG induction, the read-out will dramatically shift to the uncut profile. | ||
| - | Figure X doesn’t display a fully ‘on’ restriction profile but a gradual shift towards the restriction profile that | + | Figure X doesn’t display a fully ‘on’ restriction profile but a gradual shift towards the restriction profile that represents more uncut plasmid. This indicates that induction leads to an increase of methylation of the plasmid. Although, we still observe a gradual shift of the restriction profile meaning that the read-out doesn’t leave the intermediate state.<br\><br\> |
<h4>Log vs stationary</h4> | <h4>Log vs stationary</h4> | ||
Revision as of 13:21, 26 September 2012
 "
"