Team:Potsdam Bioware/Project/At a Glance
From 2012.igem.org
At a Glance: Antibody Generation System
Antibodies in a Nutshell
The goal of our "Antibody Generation System" is to streamline the generation and production of antibodies by integrating all steps in one cell line. Antibodies are indispensable tools for research and diagnostics and they are used in diverse applications such as ELISA, western blot, affinity purification, and imuno-histology. Antibodies also represent the most important class of biopharmaceuticals with US sales of over 18 billion US$ in 2010 (Aggarwal, 2011) and various therapeutic areas such as cancer, arthritis, and macular degeneration. Depending on the use of the antibodies specific demands must be met. In research, the murine IgG is very typical. For therapy, antibodies need to be 'humanized' to avoid immunological reactions. For some applications downsized or modified versions of antibodies can be used: such as single chain Fv fragments (scFv), Fab fragments, or heavy chain only (camelid) antibodies. However most of the time, the Fc part of the antibody provides crucial functionality like secondary antibody binding in research or cell dependent cytotoxicity in therapy. Most antibodies produced in larger scale are recombinantly expressed in CHO (chinese hamster ovary) cells, due to the fact that CHO cells mimic human glycosylation and can produce large amounts of antibodies. In general, almost 70% of all biopharmaceuticals are produced in CHO cells (CHO consortium, 2012).
State of the Art
So far, antibodies are typically produced by immunizing mice, sacrificing mice, generating hybridoma by cell fusion and selecting the desired hybridoma clones. This process is very time consuming and the expression quality varies widely. In addition, only natural murine antibodies can be obtained. For modifications or large scale production, the antibodies genes are mostly identified by phage display and then recloned in a defined expression cell line such as CHO. As this is very time consuming, E. coli based antibody fragment libraries and phage display emerged as a second route. However, in here only scFv or Fab fragments are obtained in the first place, and most applications benefit from the divalent character of the full antibody and need the Fc part. Since the Fc part requires glycosylation for stability a switch from E. coli to eukaryotic cells is necessary, which again is a laborious step.
Our Approach
We aspire to implement all steps for antibody production the in the well established CHO cells. Our method avoids animal immunization or phage display and the additional recloning and thus bears the potential to be very fast and easy to automatize from start to finish. In addition, we can implement all desired antibody formats large or small and with or without glycosilation. To accomplish our goal, we conceptually divided the steps in modules, which can be tested independently.
The Antibody Module
The design specifications for the antibody module are manifold. The antibody needs to be expressed in CHO cells in a manner that is amenable to selection and maturation as well as soluble/secreted expression. For selection we use cell surface display, which couples phenotype and genotype. Maturation is outlined in the mutations module (see below). And finally for production, we need to implement a genetic switch from cell surface expression to soluble expression. All steps require stable expression over longer periods necessitating stable transfection (i.e. chromosomal integration) of the antibody construct.
We achieved stable integration in CHO cells using the recombination based Flp-In system. As a start we used two different antibody constructs. Our "small antibody construct" gene comprises a signal peptide, a scFv directed against the EGFR (epidermal growth factor receptor), a protese celavage site, a transmembrane region and as the intracellular reporter a YFP (yellow fluorescent protein).
high affine antibodies using an antibody module, a mutation module and a selection module to ensure that the cells that express a high affine antibody survive.
For the antibody module, we transiently and stably transfected CHO cells with two antibody constructs. The first one is a single chain antibody against the epidermal growth factor receptor domain three. Additional transmembrane region and a signal peptide ensure that the construct is presented on the cell surface. The second one contains a nanobody against GFP, a Fc domain, a transmembrane region and a signal peptide that also direct the construct to the surface. A switchable region ensures the possibility of shifting from membrane standing to soluble state of the antibodies with help of Cre recombinase.
The mutation module consists of one key enzyme, the activation-induced cytidine deaminase (AID). This enzyme is commonly used in mammalian immune systems to induce the hypermuation and thus antibody maturation in activated B-lymphocytes. We created the wildtype form and a modified variant of this enzyme that has a nuclear localization sequence and no nuclear export sequence. Both of them were used for transfection into CHO cells to induce hypermutation. The transfected AID induces hypermutation in antibody transfected CHO cells and thus change the antibody binding regions stochastically.
To select CHO cells which produce high affine antibodies, we designed the selection module. This module consists of viruses which show the corresponding antigen for the nanobody GFP on the surface by using a fusion protein. The virus has an antibiotic resistance cassette. By binding the high affine antibody with the surface presenting antigen the virus is able to infect the CHO cells efficiently. Consequently, the CHO cells only survive if they produce high affine antibodies mutated by the AID.
Aggarwal S. What's fueling the biotech engine--2010 to 2011. Nat Biotechnol. 2011 Dec 8;29(12):1083-9. doi: 10.1038/nbt.2060. [http://www.ncbi.nlm.nih.gov/pubmed/22158359 PubMed PMID: 22158359.]
CHO consortium http://hugroup.cems.umn.edu/CHO
Main Results
Antibody Module: Antibody Expression
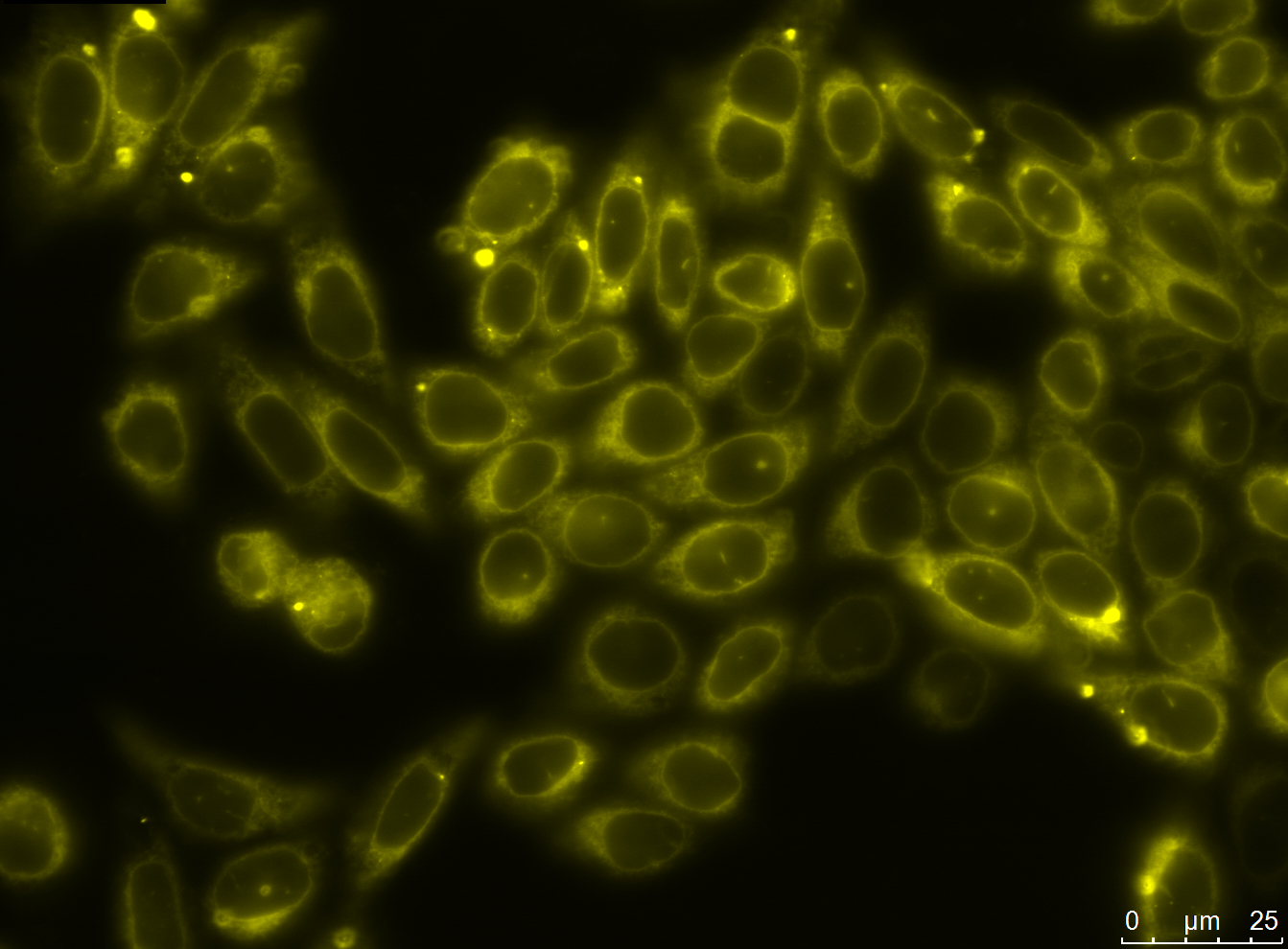
The aim of the antibody module was to design and assemble antibody constructs that would demonstrate the principle of our generation system. We designed two exemplary antibody constructs: The smaller construct contains a single chain fragment variable domain (scFv) targeting the human EGF-Receptor, a transmembrane domain, a TEV protease cleavage site and an eYFP. The advanced antibody construct consists of a nanobody binding GFP/YFP, a transmembrane domain, a TEV protease cleavage site, mCherry and two LoxP sites.
We successfully stably and transiently transfected both antibody constructs in CHO Flp-in cells. With fluorescence microscopy, confocal microscopy, immunfluorescence and FACS we were able to show the expression of the transfected parts. Under certain conditions we have seen a membrane localization of the advanced construct yet we have not been able to generate cells that transport enough molecules to their surface for successful detection. However we have shown that the switch from presenting antibodies to secreting antibodies via the Cre recombinase was successful.
Accomplished
- transient transfection of smaller antibody construct
- transient transfection of advanced antibody construct
- stable transfection of smaller construct
- stable transfection of advanced construct
- detection of expression of smaller construct
- detection of expression of advanced construct
- membrane localization of advanced construct
- proof of functionality of Cre recombinase
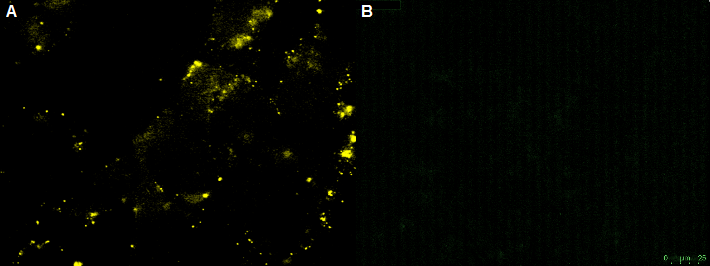
| transient transfected CHO cell lines with two differnt Antibody constructs (advanced -[http://partsregistry.org/Part:BBa_K929107 BBa_K929107]and smaller -[http://partsregistry.org/Part:BBa_K929101 BBa_K929101]) | |
| stable transfected CHO cell lines with two differnt Antibody constructs (left: advanced- [http://partsregistry.org/Part:BBa_K929107 BBa_K929107],right: smaller [http://partsregistry.org/Part:BBa_K929101 BBa_K929101]) |
Mutation Module: Antibody Maturation by Mutation with the AID Enzyme
Activation Induced Cytidine Deaminase (AID) is one of the key enzymes of antibody maturation in immune system of mammalian organisms. We used the AID to mutate the antibody sequences in CHO cells and in E. coli during Phage Display. In prior publications, it was shown that the AID is active in CHO cells and E. coli by using transfection or transformation, respectively.
The goals of the mutation module were to express the wildtype and modified AID in CHO cells, to prove the nuclear localization of the modified AID, to determine the mutation rate of wildtype AID and modified AID and to co-transfect the AID with antibody construct. The goal to maturate antibodies was not achieved.
Accomplished
- recloning of purified antibody plasmids from CHO cells in E. coli
- expression of wildtype and modified AID in CHO cells
- nuclear localization of modified AID
- determination of mutation rates of wildtype and modified AID
- co-transfection of AID and antibody construct
Selection Module: Selection or Screening for the Desired Clone
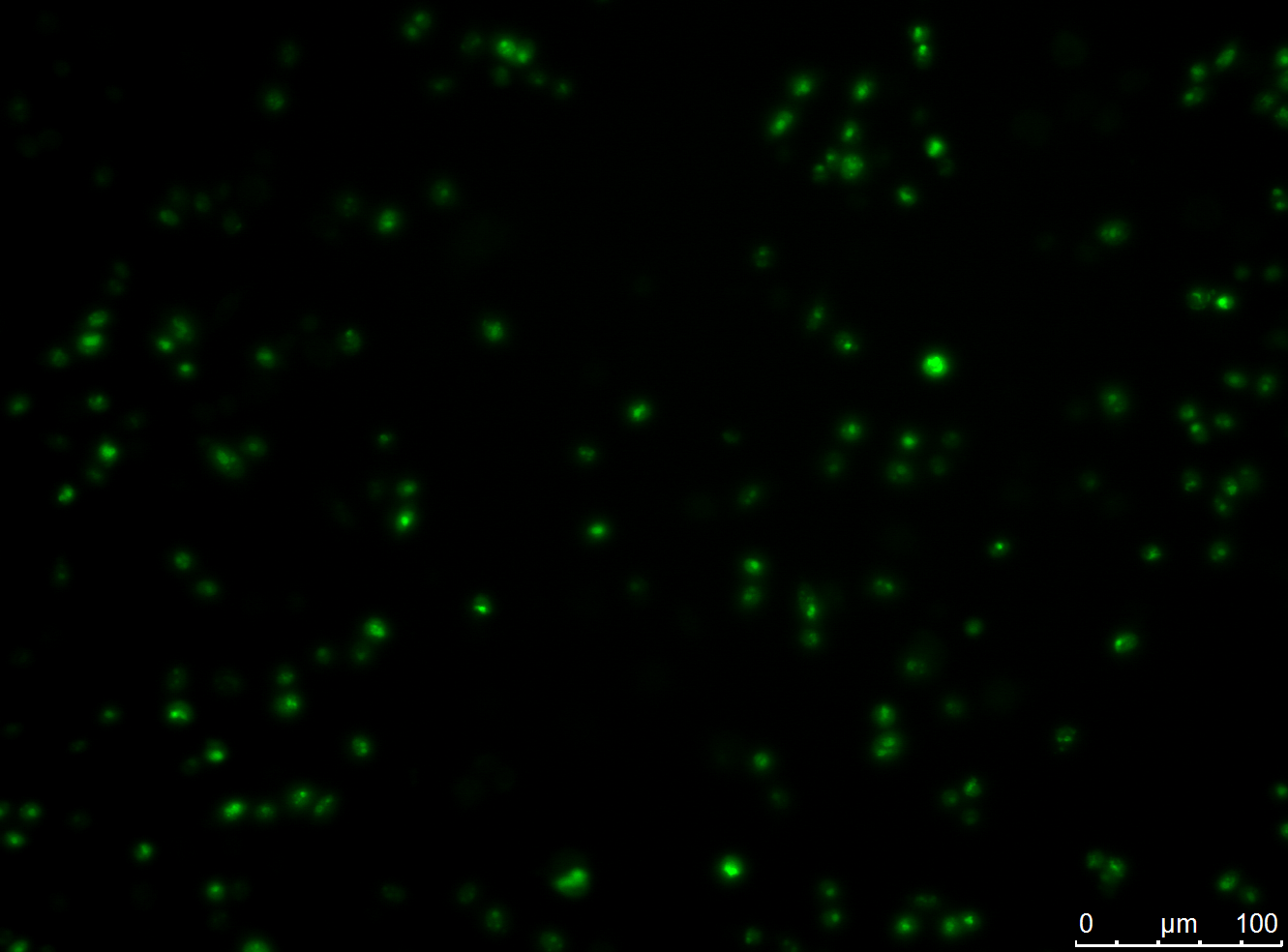
To select the desired cells from the ones expressing the antibody construct, targeted vital particles are required. Therefore, recombinant adeno-associated virus (rAAV) with yellow fluorescence protein (YFP) on the surface and cyan fluorescence protein (CFP) as Gene of interest was constructed.
HT1080 were infected with the rAAV. The rAAV with the YFP on the surface and CFP as Gene of interest was used. As the rAAV can infect the HT1080 cells, the HT1080 cells show cyan fluorescence.
CHO-cells infected with rAAV with the YFP on the surface and CFP as Gene of interest. The CHO-cells can not be infected by the rAAV, which is shown by the yellow fluorescence in the unbleached picture. Bleached CHO-cells dont show YFP fluorescence.
Sortase is an enzyme which catalyzes specific ligation of two proteins to each other. The expressed external ligand-binding 3rd domain of EGFR contains the C-terminal Sortase motif to be recognized by the enzyme Sortase. The coupled protein consisting of 3rd domain of EGFR and the Sortase motif has the size of 24.8 kDa. The N-terminal Sortase motif was fused to the N-terminus of VP2/3 gene of adeno-associated virus to allow the ligation to EGFR by the Sortase.
Modeling: Analyzing and Predicting the Viral Antibody Selection
|
|
Modeling is a powerful tool to understand complex interactions or processes. We created a model to illustrate our selection system and to get a better understanding of its behavior under different conditions. Thereby, we wanted to answer two main questions:
To answer these questions we did both deterministic and stochastic modeling using MATLAB. Furthermore, we did a huge parameter analysis for each model to check what kind of influence each parameter has on our selection system. Stochastic results showed that the selection is finished on random time points after 100 h or later. Furthermore the analysis of initial concentrations of wt cells and virus revealed that there is an optimum for initial concentrations of wt cells and virus (see Fig. X). Both to less and to excessive concentrations prevent the success of the selection system |
SocialBricks
|
The idea of SocialBricks is to divide all human practice activities into different parts: the SocialBricks. Here, the SocialBricks stand for every activity which aims to inform people about the Synthetic Biology. We hope that the term SocialBricks will be accepted like the term BioBrick and will be integrated in a registry to show the society what every team has done for more elucidation. |
Potsdam Standard - a hybrid approach to assemble your gene of interest
The main problems in using the classical approach to assemble different parts with restriction enzymes are on one hand ineffective enzymes with different optimum conditions and on the other hand illegal restriction site in the sequence which restricts the use of standard assemblies.
That is the reason why we tried to establish a new RFC with a reduced use of restriction enzymes. This cloning standard is based on the use of thiophosphate primer at the 5’ end for PCR to amplifying the insert. The insert is incubated in iodine/ethanol solution to knock out the 5’ thiophosphates. After that, the new standard cloning vector, developed by us, with a RFP expression cassette as a ligation control is digested with the enzymes Apa I and Sph I. These enzymes generate 3’ overhangs which correspond to the 3’overhang generated by knocking out the thiophosphates. The digested backbone and the pliced insert is mixed, ligated and transformed into E.coli.
To proof the new assembly standard, we insert the AID into the new standard cloning vector using the Potsdam Standard. After sequencing, we saw that the cloning was successful without any mutation in the AID sequence.
read more
 "
"