Team:Wageningen UR/ObtainingthePoleroVLP
From 2012.igem.org
(→Isolation and BioBricking of the potato leafroll virus (PLRV) coat protein.) |
(→Isolation and BioBricking of the potato leafroll virus (PLRV) coat protein.) |
||
| Line 31: | Line 31: | ||
[[File:PCR.jpg]] | [[File:PCR.jpg]] | ||
| - | Further progress will be posted asap;) | + | '''Future work''' |
| + | |||
| + | Sequencing of putative coat protein gene | ||
| + | |||
| + | Luteoviridae coat protein/readtrough analogy experiments | ||
| + | |||
| + | Transformation of the gene, inserted into iGEM pSB1C3 backbone, into E.coli | ||
| + | |||
| + | Isolation of monomers and formation of VLP’s | ||
| + | |||
| + | VLP analysis/characterisation | ||
| + | |||
| + | ''Further progress will be posted asap;)'' | ||
Revision as of 09:02, 30 July 2012
Isolation and BioBricking of the potato leafroll virus (PLRV) coat protein.
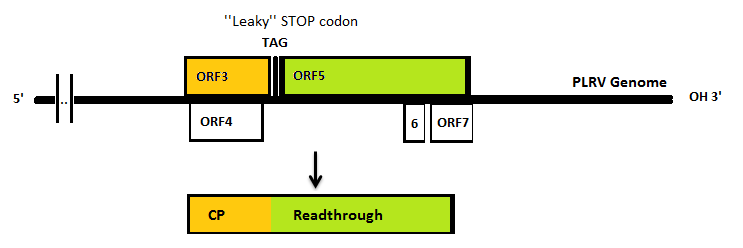
The Polero (potato leaf roll virus, PLRV) virus coat proteins can be used as building blocks to form VLP’s. PLRV is a positive sense RNA virus (group IV). Viruses from this group have their genome directly utilized as if it were mRNA. Ribosomes from the host cell translate their genome into a protein, with RNA-dependent RNA polymerase as one of it. This means that to isolate the coat protein gene we need to isolate the RNA of infected potato plants which would include the viral RNA.
After obtaining the coat protein gene of PLRV, the PCR product can be sent to be sequenced. The sequence of our isolate together with other isolates will give us information about the conserved regions in the gene. Alignment experiments can be performed, also together with data of other members of the Luteoviridae. The PLRV coat protein can thereafter be ‘’BioBricked’’. This brick can be expressed in Escherichia coli and produced monomers can be isolated and purified.
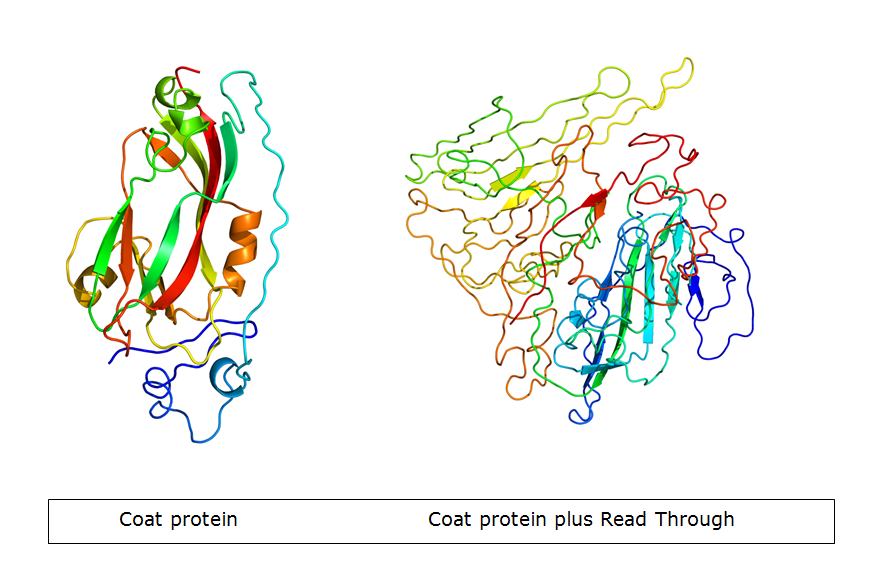
In nature, the stopcodon of the coat protein is sometimes ‘’missed’’, which results in a 70kDa readthrough product. This is considerably larger than the 23kDa coat protein on its own. When the coat protein as well as the coat protein plus readthrough are assembled together to form the virus capsid, spikes are formed on the capsid. These spike, according to literature, have a function in the transmission by aphids, primarily the green peach aphid, Myzus persicae. We expect that these spike can be changed without having a large effect on VLP formation. This, thus, seems a good strategy to build in either E-coils or K-coils, described in the general project introduction. Virus like particles (VLPs) are being formed either with or without readtrough proteins. The expression of coat protein monomers, needed for VLP formation, has never been done in E.coli. If this step succeeds, further modifications on the VLP’s will be done.
Aim: Our primary aim is to get self-assembling VLP’s from PLRV in E. coli. If we succeed, we can attempt to modify the spike on the outside.
Progress:
RNA isolation of potato leaf tissue
Reverse transcriptase reaction to obtain cDNA
PCR on cDNA using coat protein primers
Addition of Prefix and Suffix to CP genes
Future work
Sequencing of putative coat protein gene
Luteoviridae coat protein/readtrough analogy experiments
Transformation of the gene, inserted into iGEM pSB1C3 backbone, into E.coli
Isolation of monomers and formation of VLP’s
VLP analysis/characterisation
Further progress will be posted asap;)
 "
"