Team:Tianjin/Modeling/Expression
From 2012.igem.org
Austinamens (Talk | contribs) (→Modeling Objective) |
Austinamens (Talk | contribs) (→Modeling Objective) |
||
| Line 9: | Line 9: | ||
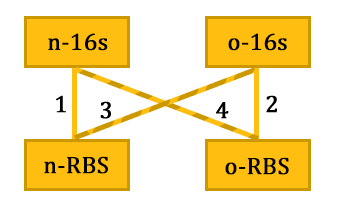
As shown in Figure 1, there are two kinds of ribosome and two mRNA with two orthogonal RBS sequence. There can be four kinds of interactions between the four components: n-16S – n-RBS, n-16S – o-RBS, o-16S – o-RBS, o-16S – n-RBS. However, there are many questions which are unclear to us and thus need answering: How can the orthogonal mRNA express their encoded protein? How can the two proteins expressed system interact with each other? Since there are too much known about the mechanism of the protein expressed with coexistence of two protein expression system. Thus, our goal of modeling is to describe the whole process with the help of mathematical tool and to predict the result of the wet lab. Specifically, there are two goals: the verification of orthogonality and the prediction of protein expression level after introducing the orthogonal system by two steps. | As shown in Figure 1, there are two kinds of ribosome and two mRNA with two orthogonal RBS sequence. There can be four kinds of interactions between the four components: n-16S – n-RBS, n-16S – o-RBS, o-16S – o-RBS, o-16S – n-RBS. However, there are many questions which are unclear to us and thus need answering: How can the orthogonal mRNA express their encoded protein? How can the two proteins expressed system interact with each other? Since there are too much known about the mechanism of the protein expressed with coexistence of two protein expression system. Thus, our goal of modeling is to describe the whole process with the help of mathematical tool and to predict the result of the wet lab. Specifically, there are two goals: the verification of orthogonality and the prediction of protein expression level after introducing the orthogonal system by two steps. | ||
| - | [[File:modeling_fig_1.png|thumb|500px|center|'''Figure 1'''.The four interactions among the four components.]] | + | [[File:modeling_fig_1.png|thumb|500px|center|'''Figure 1'''. Basic idea of our model: The four interactions among the four components.]] |
=Model Description and Design= | =Model Description and Design= | ||
Revision as of 08:25, 21 September 2012

Contents |
Background
Within our chassis bacteria, the protein expression system is special because there are two channels to express the protein: the canonical channel exists within any wild type bacteria and the orthogonal channel created artificially with the help of orthogonal ribosomes and orthogonal mRNA whose protein expression is relatively independent of the canonical ones. There are two kinds of ribosome and mRNA: host ribosome (also known as normal ribosome, canonical ribosome or n-ribosome) orthogonal-ribosome (o-ribosome), normal-mRNA (n-mRNA) with canonical RBS sequence and orthogonal mRNA (o-mRNA) with mutated RBS sequence. The canonical mRNA can be translated with the help of canonical ribosome. On the other hand, o-ribosomes can also translate genes with altered Shine-Dalgarno (SD) sequences not recognized by host ribosomes.
In the case of o-ribosomes, mutations are introduced into the ASD region such that they can base pair with complementary, noncanonical SD sequences. However, we are not familiar with the internal mechanism of how the two kinds of mRNA interact with the two kinds of ribosomes. In order to investigate the mechanism of how the four components interact with each other within our chassis bacteria, we simulate the whole process through modeling.
Modeling Objective
As shown in Figure 1, there are two kinds of ribosome and two mRNA with two orthogonal RBS sequence. There can be four kinds of interactions between the four components: n-16S – n-RBS, n-16S – o-RBS, o-16S – o-RBS, o-16S – n-RBS. However, there are many questions which are unclear to us and thus need answering: How can the orthogonal mRNA express their encoded protein? How can the two proteins expressed system interact with each other? Since there are too much known about the mechanism of the protein expressed with coexistence of two protein expression system. Thus, our goal of modeling is to describe the whole process with the help of mathematical tool and to predict the result of the wet lab. Specifically, there are two goals: the verification of orthogonality and the prediction of protein expression level after introducing the orthogonal system by two steps.
Model Description and Design
How is protein expressed with the existence of an orthogonal system?
The strength of the interaction between SD and ASD sequence is thought to influence translational efficiency as mutations in either the SD or ASD sequence that weaken the interaction reduce the amount of protein made. The mechanism of protein expression is primarily determined by the delta Gibbs free energy of the combination of SD sequence on ribosome and the ASD sequence on RBS of the mRNA. In most cases, translation initiation is the rate-limiting step. Its rate is determined by multiple molecular interactions, including the hybridization of the 16S rRNA to the RBS sequence, the binding of rRNA to the start codon, the distance between the 16S r RNA binding site and the start codon (called spacing) and the presence of RNA secondary structures that occlude either the 16S rRNA binding site or the standby site.
Where, r stands for the translation initiation rate of the protein, ∆G_tot means the total Gibbs free energy change of the SD and ASD sequence. ∆G_tot is more negative when attractive interactions between ribosome and mRNA are present, and ∆G_tot is more positive when mutually exclusive secondary structures are present. β is the apparent Boltzmann constant for the system, which converts thermodynamic free energies to temperature differences. The initiation rate is be proportional to the amount of protein expressed, as the equation (2) described
Where, the proportionality factor k_2 accounts for any ribosome-mRNA molecular interactions that are independent of mRNA sequence and any translation-independent parameters, such as the DNA copy number, the promoter’s transcription rate, the mRNA stability and the protein dilution rate.
 "
"