Team:Wageningen UR/ObtainingthePoleroVLP
From 2012.igem.org
Hanyue0731 (Talk | contribs) (→Isolation and BioBricking of the potato leaf roll virus (PLRV) coat protein.) |
Hanyue0731 (Talk | contribs) (→Potato Leaf Roll Virus(PLRS) and Turnip Yellows Virus(TuYV)) |
||
| Line 4: | Line 4: | ||
== Potato Leaf Roll Virus(PLRS) and Turnip Yellows Virus(TuYV) == | == Potato Leaf Roll Virus(PLRS) and Turnip Yellows Virus(TuYV) == | ||
<p align="justify"> | <p align="justify"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) both are members of the genus Polerovirus and family Luteoviridae, which are both positive sense RNA virus as well. (PLRV and TuYV will be called Polerovirus all together). Moreover, they both distribute all over the world and cause great yield loss for crops yearly. However, the host for PLVR and TuYV are different: PLRV mostly infect potatoes and other plants in family Solanaceae; TuTV mainly infect rapeseed (Brassica. napus) and cabbage. [1] [2] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:PLRV infected potato plants.jpg]] | |
| + | Figure 1: PLRV infected potato plants. [3] | ||
| + | The genome of PLRV and TuYV is similar to each other: both of them have a leaky coat protein stop codon. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part. It has been reported that either coat protein or coat protein with readthrough can form VLPs.[4] | ||
| - | |||
| - | [[File: | + | [[File:The PLRV genome.jpg]] |
| + | Figure 2:The PLRV genome. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[File: | + | Compared with CCMV and Hepatitis B, Polerovirus has its own unique advantage: the spike on the C terminus makes it much easier to be modified on the outside. CCMV and Hepatitis B have a loop on the outside, in order to modify them, extra extension or deletion are needed, while the C terminus of Polerovirus will be stuck out and form a spike after VLP assembly. The spike is not involved in the VLP formation, so the natural characteristics of the VLP will not be changed after modification, in this case, adding the PnAS. What’s more, the PLRV VLP has only been produced in the eukaryotic cells, more specifically, insect cells. We would like to explore the possibility to produce PLRV in prokaryotic cells, such as E.coli, which will make producing PLRV VLP less laborious and cheaper. Based on two reasons above, we choose Polerovirus. |
| + | |||
| + | [[File:Structure of PLRV coat protein.jpg]] | ||
| + | Figure 3: Structure of PLRV coat protein. | ||
| - | + | In order to show the possibility of obtaining of biobrick from nature, we asked some PLRV infected potato plant leaves from ‘Dutch General Inspection Service for Agricultural seed and seed potatoes’ (www.nak.nl) and they were sent to us by normal service. Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template. With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature, this makes the PLRV coat protein biobrick is our favorite natural biobricks. | |
| - | + | ||
| - | + | The viral gene encoding the Coat Protein for TuYV (GenBank: NC_003743.1 (3483..5495)) was obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. With designed primers, the TuYV coat protein and coat protein with partial readthrough, both with or without his-tag, were bricked into iGEM standard backbone as well. | |
| - | |||
| - | + | References: | |
| - | + | 1. Potato leafroll virus. Available from: http://en.wikipedia.org/wiki/Potato_leafroll_virus. | |
| - | + | 2. Juergens, M., et al., Genetic analyses of the host-pathogen system Turnip yellows virus (TuYV)-rapeseed (Brassica napus L.) and development of molecular markers for TuYV-resistance. Theor Appl Genet, 2010. 120(4): p. 735-44. | |
| - | + | 3. diseases: Potato leafroll luteovirus - Potato Leaf Roll Virus (PLRV) Available from: http://www.agroatlas.ru/en/content/diseases/Solani/Solani_Potato_leafroll_luteovirus/. | |
| - | + | 4. Lamb, J.W., et al., Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus. J Gen Virol, 1996. 77(Pt 7): p. 1349-58. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus | + | |
| - | + | ||
Revision as of 20:38, 20 September 2012
Potato Leaf Roll Virus(PLRS) and Turnip Yellows Virus(TuYV)
Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) both are members of the genus Polerovirus and family Luteoviridae, which are both positive sense RNA virus as well. (PLRV and TuYV will be called Polerovirus all together). Moreover, they both distribute all over the world and cause great yield loss for crops yearly. However, the host for PLVR and TuYV are different: PLRV mostly infect potatoes and other plants in family Solanaceae; TuTV mainly infect rapeseed (Brassica. napus) and cabbage. [1] [2]
 Figure 1: PLRV infected potato plants. [3]
The genome of PLRV and TuYV is similar to each other: both of them have a leaky coat protein stop codon. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part. It has been reported that either coat protein or coat protein with readthrough can form VLPs.[4]
Figure 1: PLRV infected potato plants. [3]
The genome of PLRV and TuYV is similar to each other: both of them have a leaky coat protein stop codon. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part. It has been reported that either coat protein or coat protein with readthrough can form VLPs.[4]
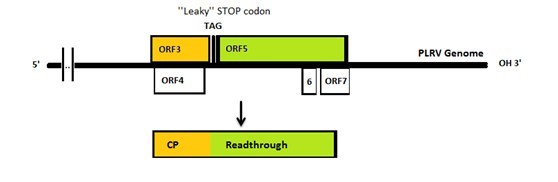 Figure 2:The PLRV genome.
Compared with CCMV and Hepatitis B, Polerovirus has its own unique advantage: the spike on the C terminus makes it much easier to be modified on the outside. CCMV and Hepatitis B have a loop on the outside, in order to modify them, extra extension or deletion are needed, while the C terminus of Polerovirus will be stuck out and form a spike after VLP assembly. The spike is not involved in the VLP formation, so the natural characteristics of the VLP will not be changed after modification, in this case, adding the PnAS. What’s more, the PLRV VLP has only been produced in the eukaryotic cells, more specifically, insect cells. We would like to explore the possibility to produce PLRV in prokaryotic cells, such as E.coli, which will make producing PLRV VLP less laborious and cheaper. Based on two reasons above, we choose Polerovirus.
Figure 2:The PLRV genome.
Compared with CCMV and Hepatitis B, Polerovirus has its own unique advantage: the spike on the C terminus makes it much easier to be modified on the outside. CCMV and Hepatitis B have a loop on the outside, in order to modify them, extra extension or deletion are needed, while the C terminus of Polerovirus will be stuck out and form a spike after VLP assembly. The spike is not involved in the VLP formation, so the natural characteristics of the VLP will not be changed after modification, in this case, adding the PnAS. What’s more, the PLRV VLP has only been produced in the eukaryotic cells, more specifically, insect cells. We would like to explore the possibility to produce PLRV in prokaryotic cells, such as E.coli, which will make producing PLRV VLP less laborious and cheaper. Based on two reasons above, we choose Polerovirus.
 Figure 3: Structure of PLRV coat protein.
In order to show the possibility of obtaining of biobrick from nature, we asked some PLRV infected potato plant leaves from ‘Dutch General Inspection Service for Agricultural seed and seed potatoes’ (www.nak.nl) and they were sent to us by normal service. Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template. With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature, this makes the PLRV coat protein biobrick is our favorite natural biobricks.
The viral gene encoding the Coat Protein for TuYV (GenBank: NC_003743.1 (3483..5495)) was obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. With designed primers, the TuYV coat protein and coat protein with partial readthrough, both with or without his-tag, were bricked into iGEM standard backbone as well.
References:
1. Potato leafroll virus. Available from: http://en.wikipedia.org/wiki/Potato_leafroll_virus.
2. Juergens, M., et al., Genetic analyses of the host-pathogen system Turnip yellows virus (TuYV)-rapeseed (Brassica napus L.) and development of molecular markers for TuYV-resistance. Theor Appl Genet, 2010. 120(4): p. 735-44.
3. diseases: Potato leafroll luteovirus - Potato Leaf Roll Virus (PLRV) Available from: http://www.agroatlas.ru/en/content/diseases/Solani/Solani_Potato_leafroll_luteovirus/.
4. Lamb, J.W., et al., Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus. J Gen Virol, 1996. 77(Pt 7): p. 1349-58.
Figure 3: Structure of PLRV coat protein.
In order to show the possibility of obtaining of biobrick from nature, we asked some PLRV infected potato plant leaves from ‘Dutch General Inspection Service for Agricultural seed and seed potatoes’ (www.nak.nl) and they were sent to us by normal service. Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template. With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature, this makes the PLRV coat protein biobrick is our favorite natural biobricks.
The viral gene encoding the Coat Protein for TuYV (GenBank: NC_003743.1 (3483..5495)) was obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. With designed primers, the TuYV coat protein and coat protein with partial readthrough, both with or without his-tag, were bricked into iGEM standard backbone as well.
References:
1. Potato leafroll virus. Available from: http://en.wikipedia.org/wiki/Potato_leafroll_virus.
2. Juergens, M., et al., Genetic analyses of the host-pathogen system Turnip yellows virus (TuYV)-rapeseed (Brassica napus L.) and development of molecular markers for TuYV-resistance. Theor Appl Genet, 2010. 120(4): p. 735-44.
3. diseases: Potato leafroll luteovirus - Potato Leaf Roll Virus (PLRV) Available from: http://www.agroatlas.ru/en/content/diseases/Solani/Solani_Potato_leafroll_luteovirus/.
4. Lamb, J.W., et al., Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus. J Gen Virol, 1996. 77(Pt 7): p. 1349-58.
 "
"









