Membrane Accelerator
As there is no compartment in prokaryotic cells, enzymes involved in a biochemical pathway diffuse all over the cytoplasm. Intermediates produced from one enzyme cannot be passed efficiently to the next due to spatial obstacles.
Rationally designed assemblies of enzymes could help substrates flow by decreasing distance that intermediates have to travel between enzymes, and thus increase yields of sequential biological reactions. If we attach those enzymes to engineered membrane proteins which constitutively assemble together, the reactions are going to proceed much faster.
Based on this theory, we tried to build a Membrane Accelerator which organizes enzymes involved in sequential reactions in order on E.coli's inner membrane.
Construction Details
As demonstrated before, we based our device on E.coli inner membrane protein Lgt. SRP-dependent signaling sequence of DsbA is fused to N-terminus to localize fusion proteins to membrane.
To rationally assemble and arrange enzymes onto inner membrane of E.coli, we incorporate three protein domains that can interact with their cognate ligands. Protein-protein interaction domains and ligands from metazoan cells (mouse SH3 and PDZ domains and rat GBD domain) were recruited to assemble enzymes on membrane. Each domain could assemble with its cognate ligand.
| File:12SJTU 2PDZ.gif
| File:12SJTU GBD.gif
| File:12SJTU SH3.gif
|
| Fig.1 : PDZ domain & ligand
| Fig.2 : GBD domain & ligand
| Fig.3 : SH3 domain & ligand
|
| from PDB 2PDZ
| from PDB 1T84
| from PDB 1CKB
|
| [http://partsregistry.org/Part:BBa_K771103 Part:BBa_K771103] and [http://partsregistry.org/Part:BBa_K771104 Part:BBa_K771104]
| [http://partsregistry.org/Part:BBa_K771105 Part:BBa_K771105] and [http://partsregistry.org/Part:BBa_K771106 Part:BBa_K771106]
| [http://partsregistry.org/Part:BBa_K771107 Part:BBa_K771107] and [http://partsregistry.org/Part:BBa_K771108 Part:BBa_K771103]
|
Recruiting interacting protein domain and ligand on fusion membrane protein could help organize each component in order.
 Fig.4 :The construction sketch of fusion membrane protein
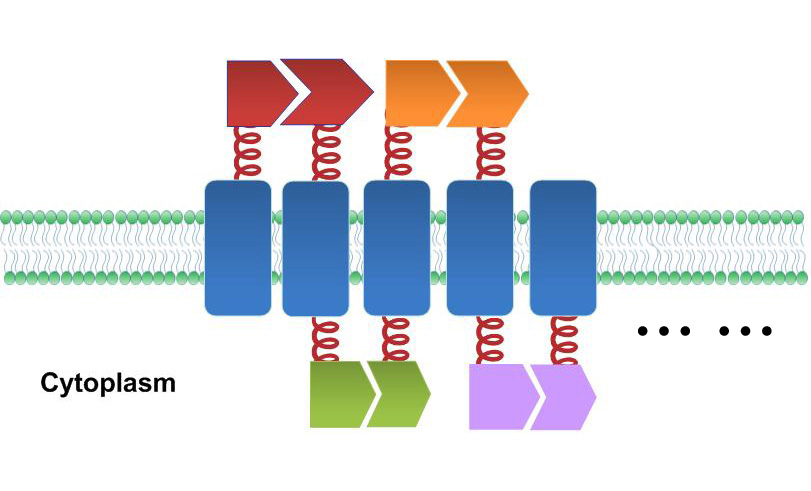 Fig.5 :Sketch of Membrane Accelerator. Membrane proteins are organized through interacting proteins. Each pair of interacting proteins is shown in the same color Fluorescence Complementation Assay
To prove that our membrane protein equipped with those interacting protein could assemble with each other, we conducted fluorescence complementation assay.
In fluorescence complementation assay, proteins that are postulated to interact are fused to unfolded complementary fragments of EGFP and expressed in E.coli. Interaction of these proteins will bring the fluorescent fragments within proximity, allowing the reporter protein to reform in its native three-dimensional structure and emit its fluorescent signal. We have constructed two split EGFP domains, [http://partsregistry.org/Part:BBa_K771113 Part:BBa_K771113] and [http://partsregistry.org/Part:BBa_K771114 Part:BBa_K771114], respectively for 1EGFP and 2EGFP.
EGFP was split into two halves, named 1EGFP and 2EGFP respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed.
The membrane proteins, which we call Membrane Anchors, are registered as [http://partsregistry.org/Part:BBa_K771002 Part:BBa_K771002], [http://partsregistry.org/Part:BBa_K771003 Part:BBa_K771003], [http://partsregistry.org/Part:BBa_K771004 Part:BBa_K771004],[http://partsregistry.org/Part:BBa_K771005 Part:BBa_K771005], named Membrane Anchor 2 without MS2, Membrane Anchor 2,Membrane Anchor 3, Membrane Anchor 4 without VVD respectively.
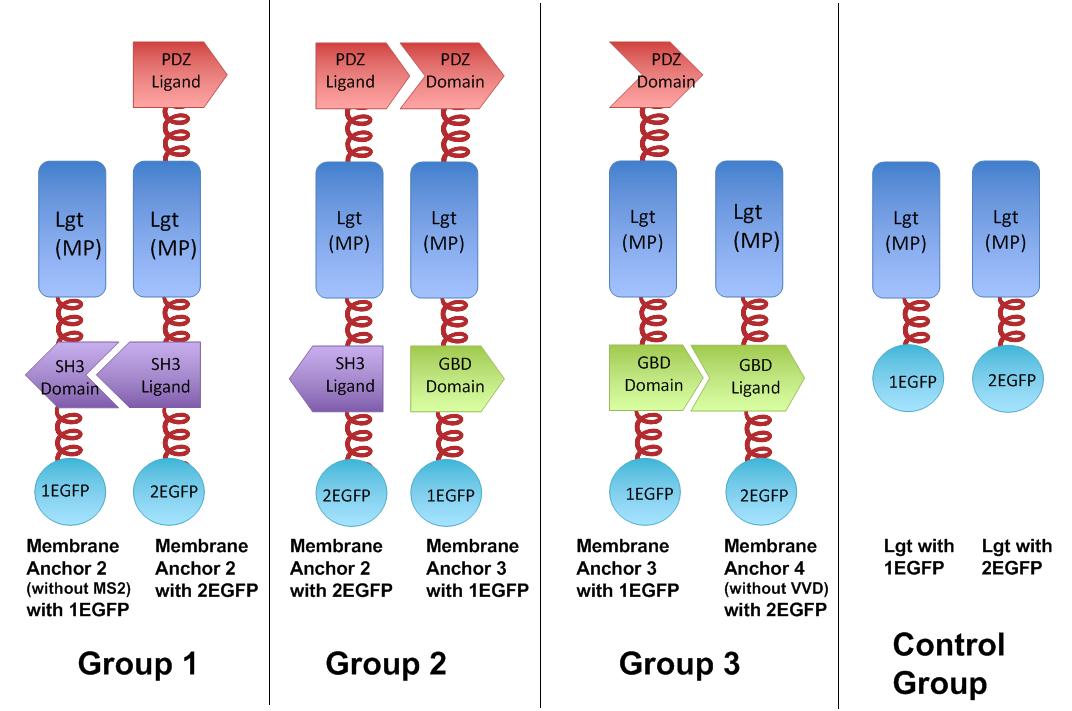 Fig.6 :Group 1, 2, 3 are designed to test the interaction of SH3domain & ligand, PDZdomain & ligand and GBDdomain & ligand respectively. Proteins within each group were coexpressed in E.coli. If there exists interaction within membrane proteins of each group, we expected to observe green fluorescence in E.coli. Control group is expected to show much weaker fluorescence. Proteins within each group are coexpressed in E.coli. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6 hours. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope.
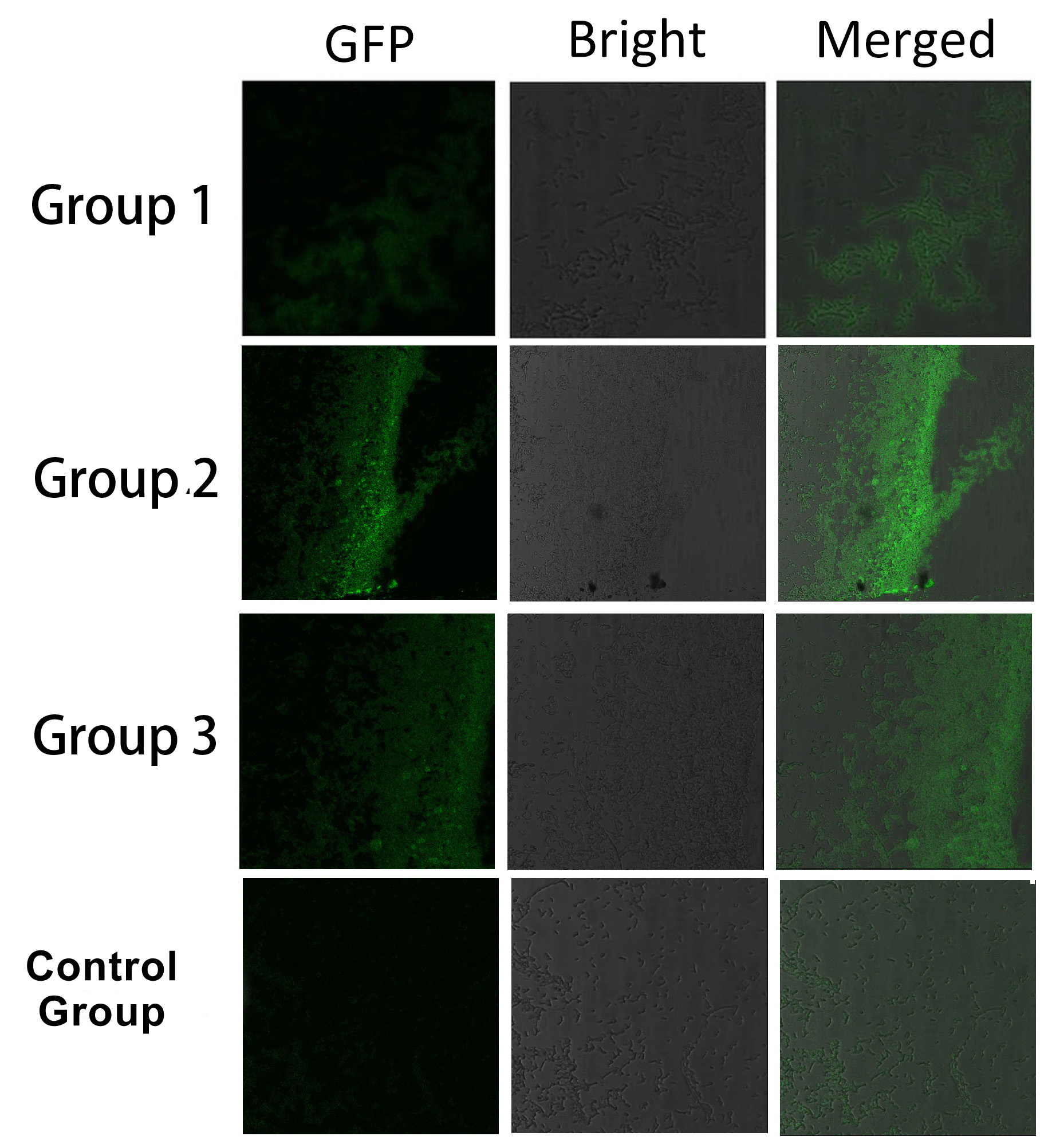 Fig.7 :Bacteria in Group 1, 2 and 3 all show significantly stronger fluorescence intensity than Control Group. So we can conclude that through recruiting interacting domain and ligand, we can assemble proteins into a complex. It is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting E.coli native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a Membrane Accelerator.
The construction of Membrane Anchors with split EGFP are [http://partsregistry.org/Part:BBa_K771402 Part:BBa_K771402], [http://partsregistry.org/Part:BBa_K771403 Part:BBa_K771403], [http://partsregistry.org/Part:BBa_K771404 Part:BBa_K771404], [http://partsregistry.org/Part:BBa_K771405 Part:BBa_K771405], respectively.
Application
Membrane Accelerator could be easily put into use by attaching functional enzymes to corresponding membrane anchors.
To verify the superiority of membrane accelerator system, we recruited Fatty Acids Biosynthetic pathway, and proved that biosynthesis rate can be increased sharply with Membrane Accelerator.
What's more, we noticed that previously published researches on Scaffold system all focused on accelerating Biosynthetic pathway, which would no doubt have practical significance. Nevertheless, no one has before applied scaffold system into Biodegradation. Biodegradation plays crucial part in environment restoration, especially at such a time when pollution is more serious than ever. Some Polycyclic Aromatic Hydrocarbons are toxic, mutagenic, carcinogenic and persist very long in environment. Sulfur in crude oil could cause various problems. Thus, we are trying to recruit Membrane Accelerator to accelerate PAH degradation and DBT desulfurization pathway.
Reference
1.Dueber, J. E., G. C. Wu, et al. (2009). "Synthetic protein scaffolds provide modular control over metabolic flux." Nature biotechnology 27(8): 753-759.
2.Valencia-Burton, M., R. M. McCullough, et al. (2007). "RNA visualization in live bacterial cells using fluorescent protein complementation." Nature Methods 4(5): 421-427.
|  "
"



