Team:Wageningen UR/Journal/week14
From 2012.igem.org
(→Lab work) |
(→Lab work) |
||
| (14 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
== Lab work == | == Lab work == | ||
| - | ''' | + | '''General''' |
| - | + | PCR st.10 Berkeley KILR | |
| - | ''30.July'' | + | Follow up of the experiment started in week 9 to switch the BBa_K197022 into a iGEM standard 10 compatible pSB1C3 backbone. Two self-designed primers were used to amplify the KILR construct out of the original BBa_K197038 backbone . |
| + | |||
| + | *FW primer Berkeley KILR: 5’ GTTTCTTCGAATTCGCGGCCGCTTCTAGAGGATCTGTTAAAGAACTGGAAGACAAAAA 3’ | ||
| + | *BW primer Berkeley KILR: 5’ TACTAGTAGCGGCCGCTGCAGGAAGAAACCGCAACCACCACGTTCAC 3’ | ||
| + | |||
| + | PCR purification of the two samples with a kit of Thermo Scientific (yield: ca. 40 ng/µl) | ||
| + | |||
| + | [[File:Robert KILR PCR 3-8.jpg|500px|thumb|center|''Figure 1: PCR standard 10 Berkeley KILR'']] | ||
| + | |||
| + | |||
| + | digestion PCR product Berkeley coil | ||
| + | |||
| + | Prerequired step for the final ligation into pSB1C3. Both donor backbone (BBa_J04450) and the PCR product were digested with the same approach by using the restriction enzymes EcoRI and PstI. Using PstI requires another PCR purification (yield: ca.15 ng/ µl) before ligation and transformation. | ||
| + | |||
| + | ''' CCMV ''' | ||
| + | |||
| + | ''30th July'' (Hugo) | ||
| + | <br/> | ||
| + | Repeated the PCR reaction of the second PCR at 27-07. Results were still not satisfying, so had to repeat the reaction even another time. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''31st July'' (Hugo) | ||
| + | <br/> | ||
| + | PCR product purification of second PCR of 30-07. | ||
| + | <br/> | ||
| + | Another repeat of second PCR at 27-07. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''1st August'' (Hugo & Jeroen) | ||
| + | <br/> | ||
| + | Gel purification of PCR product of 31-07. | ||
| + | |||
| + | Another repeat of second PCR at 27-07, now with annealing temperature of 58 °C. | ||
| + | <br/> | ||
| + | Marnix did another repeat of the second PCR at 27-07, now with an annealing temperature of 59 °C. Next to that, Marnix uses only 1 uL of each primer. From here onwards, I myself also use only 1 uL of each primer. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Colony pcr from plates containing transformed DH5alpha with all CCMV fragments. because all fragments are compatible with the delta 25 primers, these are used in the master mix. however this will give the same size products | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Pcr1-8.PNG|500px|thumb|center|''Figure 2: colony PCR'']] | ||
| + | |||
| + | |||
| + | 1 till 10 are CCMV products | ||
| + | 11 till 15 are his CCMV products | ||
| + | 16 till 20 are delta 25 CCMV products | ||
| + | 21 till 25 are delta 25 his CCMV products | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''2nd August'' (Hugo & Jeroen) | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Another repeat of second PCR at 27-07. Annealing temperature of 59 °C. This time also with dreamTaq. It appeared that both Phusion and DreamTaq negative controls have a band (positive negative). | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Digestion Check of the grown plasmids gives that CCMV and delta 25 fragments of yesterday indeed where of the right size. plasmids of the HIS CCMV and delta 25 his did not confirm | ||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | ''3rd August'' (Hugo) | ||
| + | <br/> | ||
| + | <br/> | ||
| + | PCR purification of amplicon of Marnix’s PCR on 01-08 followed by digestion with PstI + EcoRI. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Started the combination of IPTG induction brick with the delta 25 His CCMV and HIS CCMV pcr products to instantly make inducable parts. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Digestion of the brick with SpeI and PstI, PCR products with XbaI and PstI | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Inactivation of the restriction enzymes by PCR purification column | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Ligation for 1 hour | ||
| + | <br/> | ||
| + | <br/> | ||
| + | Transformation into DH5alpha | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ''4th August'' (Hugo) | ||
| + | <br/> | ||
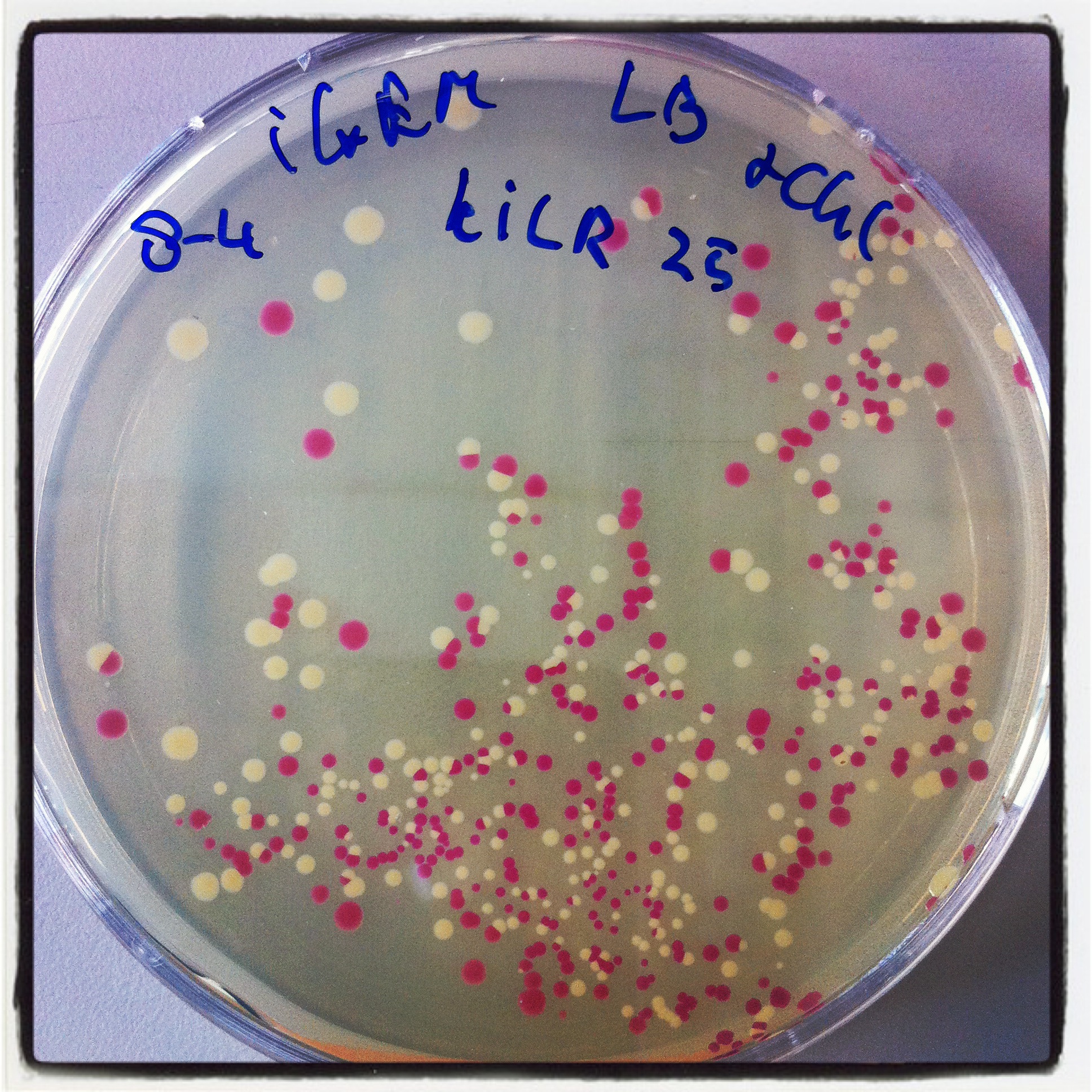
| + | Robert did ligation, followed by transformation. Result was similar to his transformants: | ||
| + | |||
| + | |||
| + | [[File:Nicetransf.JPG|500px|thumb|center|''Figure 3: Robert's red and white transformants'']] | ||
| + | |||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | ''' Hepatitis B ''' | ||
| + | |||
| + | ''30. July'' | ||
* Miniprepping of successful transformants from the transformation (Hep B + BBa_J04500) | * Miniprepping of successful transformants from the transformation (Hep B + BBa_J04500) | ||
| Line 16: | Line 109: | ||
| - | ''1.August'' | + | ''1. August'' |
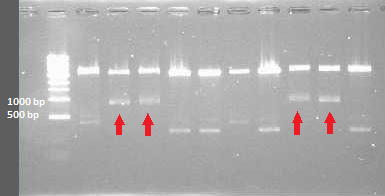
-> the digestion check was repeated 2 times (once with a longer incubation time and once with a higher amount of DNA in the digestion mixture) until the insert size could be seen on the gel | -> the digestion check was repeated 2 times (once with a longer incubation time and once with a higher amount of DNA in the digestion mixture) until the insert size could be seen on the gel | ||
| - | [[File:digestion check 1.August.png| | + | [[File:digestion check 1.August.png|300px|thumb|center|''Figure 4: digestion check 1.August'']] |
| + | |||
| - | |||
-> the digestion check confirmed the right insert sizes in the samples they where expected (around 800bp) | -> the digestion check confirmed the right insert sizes in the samples they where expected (around 800bp) | ||
| Line 34: | Line 127: | ||
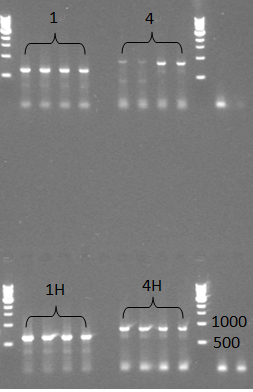
* We did two colony PCR to check the samples, but we got negative results from the colony PCR. Even so, we still continued to miniprep those colonoes which were used for colony PCR and checked them with normal PCR later, this time we saw expected bands for all 4 samples, which should be around 600bp for 1(TuYV coat protein) and 1H(TuYV coat protein with his-tag) and 800bp for 4(TuYV coat protein with partial readthrough) and 4H (TuYV coat protein with partial readthrough with his-tag). | * We did two colony PCR to check the samples, but we got negative results from the colony PCR. Even so, we still continued to miniprep those colonoes which were used for colony PCR and checked them with normal PCR later, this time we saw expected bands for all 4 samples, which should be around 600bp for 1(TuYV coat protein) and 1H(TuYV coat protein with his-tag) and 800bp for 4(TuYV coat protein with partial readthrough) and 4H (TuYV coat protein with partial readthrough with his-tag). | ||
| - | [[File:PCR check for miniprep products (TuYV in ampicillin resistant backbone).jpg]] | + | [[File:PCR check for miniprep products (TuYV in ampicillin resistant backbone).jpg|500px|thumb|center|''Figure 5: PCR check for the TuYV constructs 1, 1H 4, and 4H in the pSB1A3 backbone'' ]] |
'''PLRV''' | '''PLRV''' | ||
| - | + | <p align="justify"> | |
The PCR products obtained after Reverse Transcription followed by PCR were used as template to add iGEM prefix and suffix. The products were inserted into the backbone with Ampicilin/Kanamycin resistance. | The PCR products obtained after Reverse Transcription followed by PCR were used as template to add iGEM prefix and suffix. The products were inserted into the backbone with Ampicilin/Kanamycin resistance. | ||
| - | + | <br><br> | |
Also the PLRV coat protein plus read trough (PLRV CP_RT) was obtained by PCR reactions. We wanted to add iGEM prefix and suffix to that gene but after successful addition we found out that there are two illegal restriction sites in the read through part. This was concluded from the DNA sequences available in the GenBank database. | Also the PLRV coat protein plus read trough (PLRV CP_RT) was obtained by PCR reactions. We wanted to add iGEM prefix and suffix to that gene but after successful addition we found out that there are two illegal restriction sites in the read through part. This was concluded from the DNA sequences available in the GenBank database. | ||
| - | + | </p> | |
---- | ---- | ||
[[https://2012.igem.org/Team:Wageningen_UR/Journal/week13 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week15 next week]] | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week13 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week15 next week]] | ||
Latest revision as of 03:30, 27 September 2012
week 14: 30 july - 5 august
Lab work
General
PCR st.10 Berkeley KILR
Follow up of the experiment started in week 9 to switch the BBa_K197022 into a iGEM standard 10 compatible pSB1C3 backbone. Two self-designed primers were used to amplify the KILR construct out of the original BBa_K197038 backbone .
- FW primer Berkeley KILR: 5’ GTTTCTTCGAATTCGCGGCCGCTTCTAGAGGATCTGTTAAAGAACTGGAAGACAAAAA 3’
- BW primer Berkeley KILR: 5’ TACTAGTAGCGGCCGCTGCAGGAAGAAACCGCAACCACCACGTTCAC 3’
PCR purification of the two samples with a kit of Thermo Scientific (yield: ca. 40 ng/µl)
digestion PCR product Berkeley coil
Prerequired step for the final ligation into pSB1C3. Both donor backbone (BBa_J04450) and the PCR product were digested with the same approach by using the restriction enzymes EcoRI and PstI. Using PstI requires another PCR purification (yield: ca.15 ng/ µl) before ligation and transformation.
CCMV
30th July (Hugo)
Repeated the PCR reaction of the second PCR at 27-07. Results were still not satisfying, so had to repeat the reaction even another time.
31st July (Hugo)
PCR product purification of second PCR of 30-07.
Another repeat of second PCR at 27-07.
1st August (Hugo & Jeroen)
Gel purification of PCR product of 31-07.
Another repeat of second PCR at 27-07, now with annealing temperature of 58 °C.
Marnix did another repeat of the second PCR at 27-07, now with an annealing temperature of 59 °C. Next to that, Marnix uses only 1 uL of each primer. From here onwards, I myself also use only 1 uL of each primer.
Colony pcr from plates containing transformed DH5alpha with all CCMV fragments. because all fragments are compatible with the delta 25 primers, these are used in the master mix. however this will give the same size products
1 till 10 are CCMV products
11 till 15 are his CCMV products
16 till 20 are delta 25 CCMV products
21 till 25 are delta 25 his CCMV products
2nd August (Hugo & Jeroen)
Another repeat of second PCR at 27-07. Annealing temperature of 59 °C. This time also with dreamTaq. It appeared that both Phusion and DreamTaq negative controls have a band (positive negative).
Digestion Check of the grown plasmids gives that CCMV and delta 25 fragments of yesterday indeed where of the right size. plasmids of the HIS CCMV and delta 25 his did not confirm
3rd August (Hugo)
PCR purification of amplicon of Marnix’s PCR on 01-08 followed by digestion with PstI + EcoRI.
Started the combination of IPTG induction brick with the delta 25 His CCMV and HIS CCMV pcr products to instantly make inducable parts.
Digestion of the brick with SpeI and PstI, PCR products with XbaI and PstI
Inactivation of the restriction enzymes by PCR purification column
Ligation for 1 hour
Transformation into DH5alpha
4th August (Hugo)
Robert did ligation, followed by transformation. Result was similar to his transformants:
Hepatitis B
30. July
- Miniprepping of successful transformants from the transformation (Hep B + BBa_J04500)
- Digestion check of those minipreps
1. August
-> the digestion check was repeated 2 times (once with a longer incubation time and once with a higher amount of DNA in the digestion mixture) until the insert size could be seen on the gel
-> the digestion check confirmed the right insert sizes in the samples they where expected (around 800bp)
TuYV
- The four 'brickable' TuYV CP gene amplicons (1,1H,4,4H) were digested, ligated into the backbone from BBa_J04450, which is ampicillin resistant and transformed into our electrocompetent Mach1 E coli.
- Colony check: We saw many colonies on all 4 sample plates this time.
- We did two colony PCR to check the samples, but we got negative results from the colony PCR. Even so, we still continued to miniprep those colonoes which were used for colony PCR and checked them with normal PCR later, this time we saw expected bands for all 4 samples, which should be around 600bp for 1(TuYV coat protein) and 1H(TuYV coat protein with his-tag) and 800bp for 4(TuYV coat protein with partial readthrough) and 4H (TuYV coat protein with partial readthrough with his-tag).
PLRV
The PCR products obtained after Reverse Transcription followed by PCR were used as template to add iGEM prefix and suffix. The products were inserted into the backbone with Ampicilin/Kanamycin resistance.
Also the PLRV coat protein plus read trough (PLRV CP_RT) was obtained by PCR reactions. We wanted to add iGEM prefix and suffix to that gene but after successful addition we found out that there are two illegal restriction sites in the read through part. This was concluded from the DNA sequences available in the GenBank database.
 "
"