Team:LMU-Munich/Project
From 2012.igem.org
(→Stop Germination) |
Franzi.Duerr (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== '''Overall project''' == | == '''Overall project''' == | ||
| Line 60: | Line 40: | ||
=== Fuse Spore Coat Proteins === | === Fuse Spore Coat Proteins === | ||
| - | There are several different coat proteins in the spores of Bacillus subtilis and we chose CotZ and CgeA which are located in the outermost layer. The first step is to fuse GFP to these proteins to see if they appear on the surface and if there is any affection of spore formation. The next step is to fuse proteins with special features | + | There are several different coat proteins in the spores of Bacillus subtilis and we chose CotZ and CgeA which are located in the outermost layer. The first step is to fuse GFP to these proteins to see if they appear on the surface and if there is any affection of spore formation. The next step is to fuse proteins with special features to CotZ and CgeA to produce functional SporoBeads. |
| + | These properties could be: | ||
| + | - binding harmful viruses | ||
| + | - binding toxic metals | ||
| + | - binding plastic molecules | ||
| + | - expose enzymes | ||
| + | to filter fluids or for use in the laboratory. | ||
=== Stop Germination === | === Stop Germination === | ||
Revision as of 12:49, 23 April 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Overall project
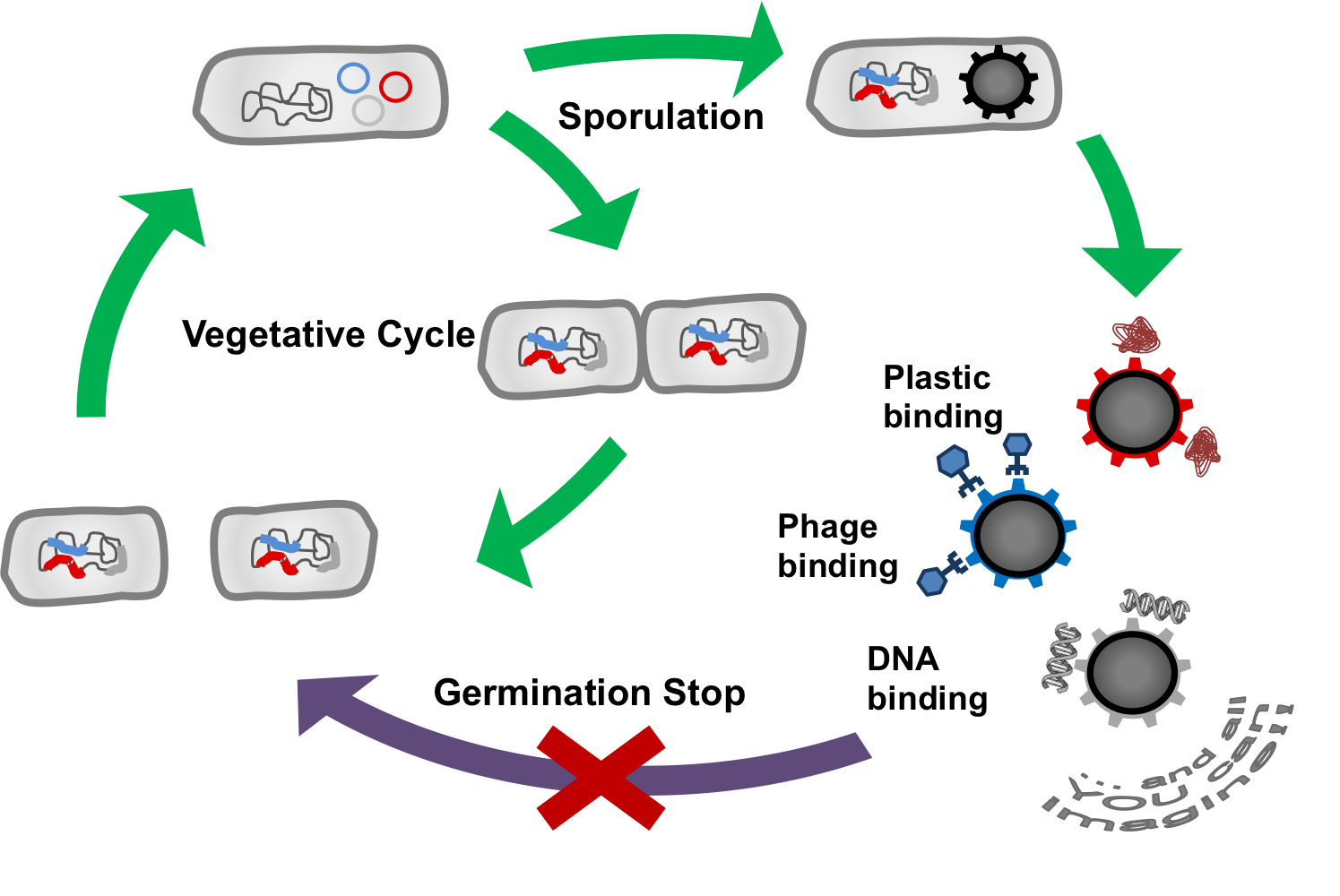
The goal of our project BEADzillus is the production of Bacillus subtilis spores which display different proteins with special features on their surface. For example, the spore could:
- bind harmful viruses
- bind toxic metals
- bind plastic molecules
- expose enzymes
to filter fluids or for use in laboratory.
As a safety issue we will stop spores from outgrowing. First we want to knock out several genes which are important for germination and second we want potentially outgrowing spores to be killed by a toxin they produce themselves.
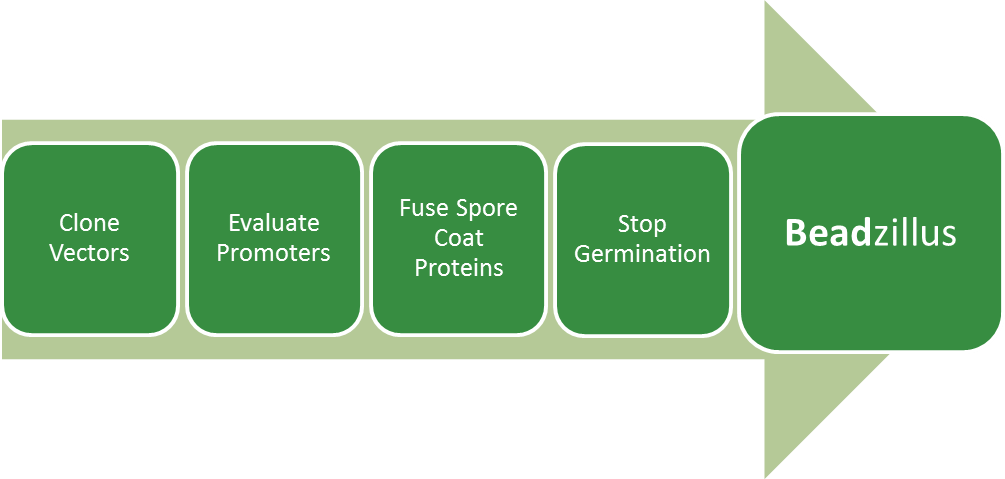
Project Details
Clone Vectors
Part of establishing Bacillus subtilis to the BioBrick mode is to modify standard vectors of Bacillus.
Evaluate Promoters
Another point is to produce promoters of Bacillus subtilis in BioBrick mode and to evaluate them.
Fuse Spore Coat Proteins
There are several different coat proteins in the spores of Bacillus subtilis and we chose CotZ and CgeA which are located in the outermost layer. The first step is to fuse GFP to these proteins to see if they appear on the surface and if there is any affection of spore formation. The next step is to fuse proteins with special features to CotZ and CgeA to produce functional SporoBeads. These properties could be: - binding harmful viruses - binding toxic metals - binding plastic molecules - expose enzymes to filter fluids or for use in the laboratory.
Stop Germination
To stop the „SporoBeads“ from germination, we thought of two different manipulations. First, we knock-out several genes which are necessary for all germination pathways, namely the cortex hydrolysis. Several gene deletion combinations will be tested for their outgrowth rates.
As a second mechanism, spores which begin to germinate despite the gene deletions shall be killed by an endotoxin system. Here, we work with a sigma-factor from a different species which works in Bacillus subtilis but does not interfere with endogenous promoters. This sigma factor is placed downstream of a promoter that is strongly activated by sigmaG , the last active sigma factor in the endospore. Therefore, the alternative sigma factor is produced right before sporulation. The bacteria also have a second construct inserted into their genome: a promoter activated by the alternative sigma factor which leads to the transcription of an endotoxin. So, if a spore starts metabolism, the toxin gene will be activated and kill the outgrowing spore.
Results
 "
"